Zinc »
PDB 7doy-7dws »
7ds1 »
Zinc in PDB 7ds1: Crystal Structure of Aspergillus Oryzae RIB2 Deaminase in Complex with Daripp (C-Terminal Deletion Mutant at pH 6.5)
Protein crystallography data
The structure of Crystal Structure of Aspergillus Oryzae RIB2 Deaminase in Complex with Daripp (C-Terminal Deletion Mutant at pH 6.5), PDB code: 7ds1
was solved by
S.C.Chen,
S.H.Liaw,
C.H.Hsu,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 31.54 / 1.58 |
| Space group | P 21 21 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 59.32, 90.778, 40.833, 90, 90, 90 |
| R / Rfree (%) | 18.3 / 20.8 |
Zinc Binding Sites:
The binding sites of Zinc atom in the Crystal Structure of Aspergillus Oryzae RIB2 Deaminase in Complex with Daripp (C-Terminal Deletion Mutant at pH 6.5)
(pdb code 7ds1). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total only one binding site of Zinc was determined in the Crystal Structure of Aspergillus Oryzae RIB2 Deaminase in Complex with Daripp (C-Terminal Deletion Mutant at pH 6.5), PDB code: 7ds1:
In total only one binding site of Zinc was determined in the Crystal Structure of Aspergillus Oryzae RIB2 Deaminase in Complex with Daripp (C-Terminal Deletion Mutant at pH 6.5), PDB code: 7ds1:
Zinc binding site 1 out of 1 in 7ds1
Go back to
Zinc binding site 1 out
of 1 in the Crystal Structure of Aspergillus Oryzae RIB2 Deaminase in Complex with Daripp (C-Terminal Deletion Mutant at pH 6.5)
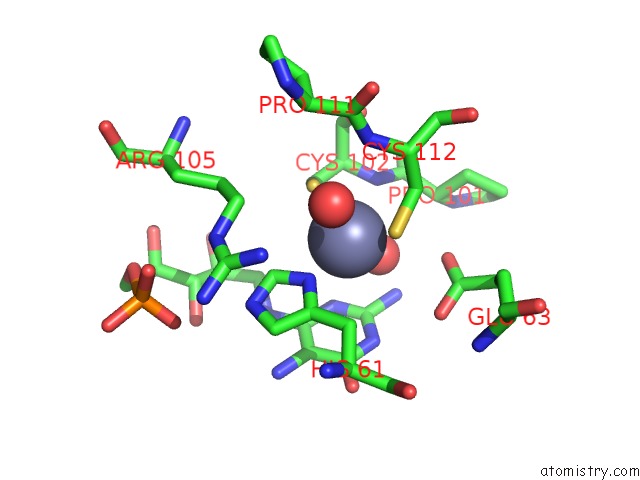
Mono view
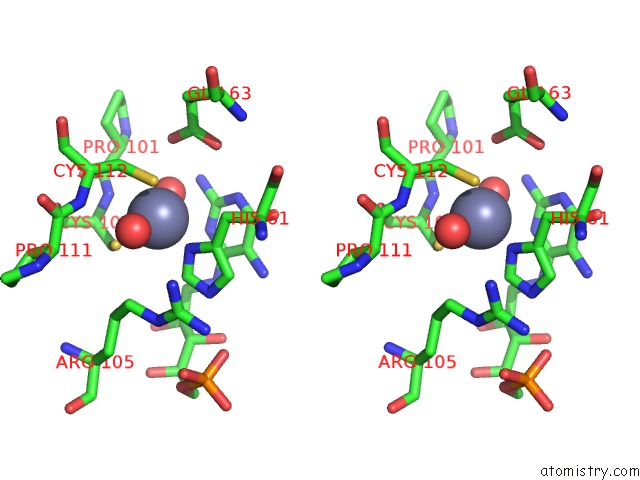
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Crystal Structure of Aspergillus Oryzae RIB2 Deaminase in Complex with Daripp (C-Terminal Deletion Mutant at pH 6.5) within 5.0Å range:
|
Reference:
S.C.Chen,
L.C.Ye,
T.M.Yen,
R.X.Zhu,
C.Y.Li,
S.C.Chang,
S.H.Liaw,
C.H.Hsu.
Crystal Structures of Aspergillus Oryzae RIB2 Deaminase: the Functional Mechanism Involved in Riboflavin Biosynthesis. Iucrj V. 8 549 2021.
ISSN: ESSN 2052-2525
DOI: 10.1107/S205225252100275X
Page generated: Tue Oct 29 19:15:19 2024
ISSN: ESSN 2052-2525
DOI: 10.1107/S205225252100275X
Last articles
Mg in 5GTIMg in 5GTH
Mg in 5H1Y
Mg in 5H1B
Mg in 5H1C
Mg in 5H0V
Mg in 5GZA
Mg in 5GZ9
Mg in 5GYN
Mg in 5GXV