Zinc »
PDB 7d52-7ddp »
7d7c »
Zinc in PDB 7d7c: Cryoem Structure of GP55-Dependent Rna Polymerase-Promoter Open Complex
Enzymatic activity of Cryoem Structure of GP55-Dependent Rna Polymerase-Promoter Open Complex
All present enzymatic activity of Cryoem Structure of GP55-Dependent Rna Polymerase-Promoter Open Complex:
2.7.7.6;
2.7.7.6;
Other elements in 7d7c:
The structure of Cryoem Structure of GP55-Dependent Rna Polymerase-Promoter Open Complex also contains other interesting chemical elements:
| Magnesium | (Mg) | 1 atom |
Zinc Binding Sites:
The binding sites of Zinc atom in the Cryoem Structure of GP55-Dependent Rna Polymerase-Promoter Open Complex
(pdb code 7d7c). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total 2 binding sites of Zinc where determined in the Cryoem Structure of GP55-Dependent Rna Polymerase-Promoter Open Complex, PDB code: 7d7c:
Jump to Zinc binding site number: 1; 2;
In total 2 binding sites of Zinc where determined in the Cryoem Structure of GP55-Dependent Rna Polymerase-Promoter Open Complex, PDB code: 7d7c:
Jump to Zinc binding site number: 1; 2;
Zinc binding site 1 out of 2 in 7d7c
Go back to
Zinc binding site 1 out
of 2 in the Cryoem Structure of GP55-Dependent Rna Polymerase-Promoter Open Complex
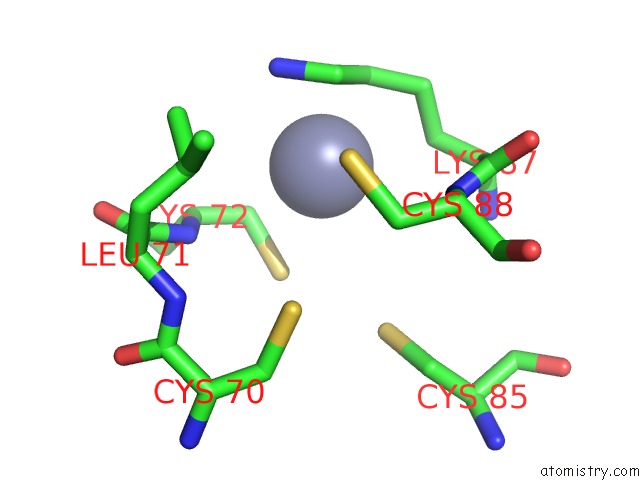
Mono view
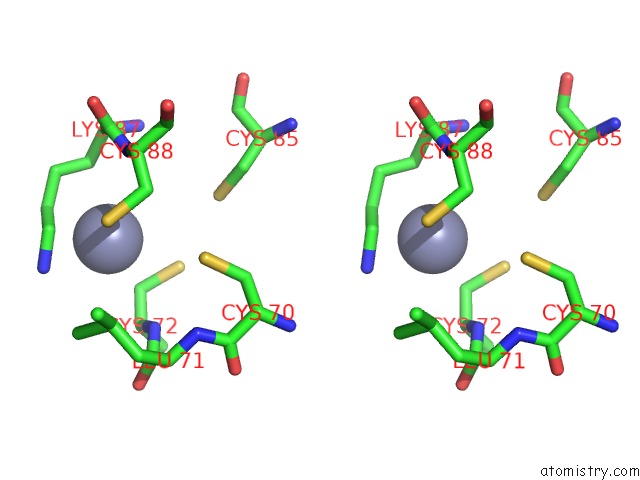
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Cryoem Structure of GP55-Dependent Rna Polymerase-Promoter Open Complex within 5.0Å range:
|
Zinc binding site 2 out of 2 in 7d7c
Go back to
Zinc binding site 2 out
of 2 in the Cryoem Structure of GP55-Dependent Rna Polymerase-Promoter Open Complex
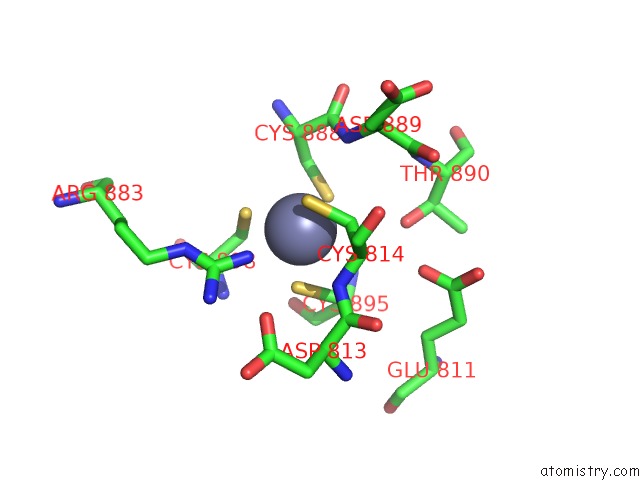
Mono view
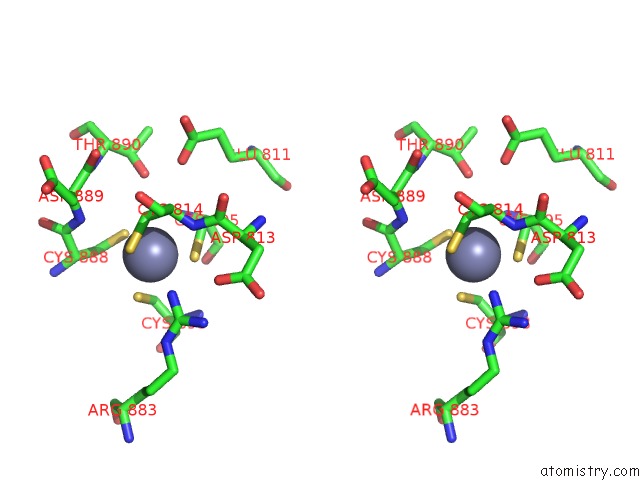
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 2 of Cryoem Structure of GP55-Dependent Rna Polymerase-Promoter Open Complex within 5.0Å range:
|
Reference:
J.Shi,
A.Wen,
S.Jin,
B.Gao,
Y.Huang,
Y.Feng.
Transcription Activation By A Sliding Clamp. Nat Commun 2021.
ISSN: ESSN 2041-1723
Page generated: Tue Oct 29 18:44:44 2024
ISSN: ESSN 2041-1723
Last articles
Mg in 1YITMg in 1YIJ
Mg in 1YJF
Mg in 1YI2
Mg in 1YJ2
Mg in 1YHQ
Mg in 1YIO
Mg in 1YHM
Mg in 1YID
Mg in 1YI1