Zinc »
PDB 6yzr-6z9b »
6z3a »
Zinc in PDB 6z3a: MEC1-DDC2 (Wild-Type) in Complex with Amp-Pnp
Enzymatic activity of MEC1-DDC2 (Wild-Type) in Complex with Amp-Pnp
All present enzymatic activity of MEC1-DDC2 (Wild-Type) in Complex with Amp-Pnp:
2.7.11.1;
2.7.11.1;
Zinc Binding Sites:
The binding sites of Zinc atom in the MEC1-DDC2 (Wild-Type) in Complex with Amp-Pnp
(pdb code 6z3a). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total 2 binding sites of Zinc where determined in the MEC1-DDC2 (Wild-Type) in Complex with Amp-Pnp, PDB code: 6z3a:
Jump to Zinc binding site number: 1; 2;
In total 2 binding sites of Zinc where determined in the MEC1-DDC2 (Wild-Type) in Complex with Amp-Pnp, PDB code: 6z3a:
Jump to Zinc binding site number: 1; 2;
Zinc binding site 1 out of 2 in 6z3a
Go back to
Zinc binding site 1 out
of 2 in the MEC1-DDC2 (Wild-Type) in Complex with Amp-Pnp
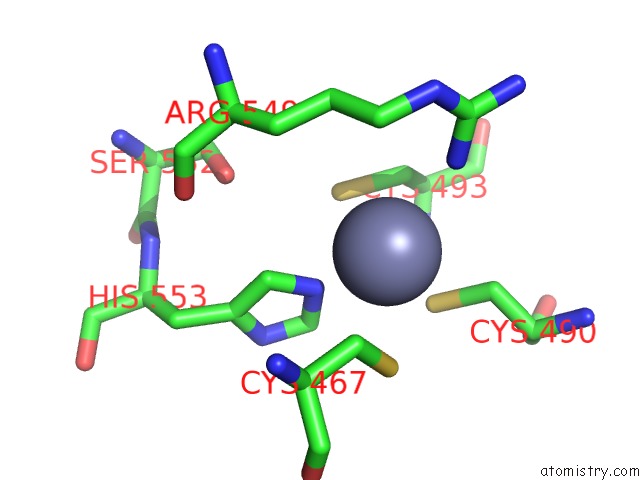
Mono view
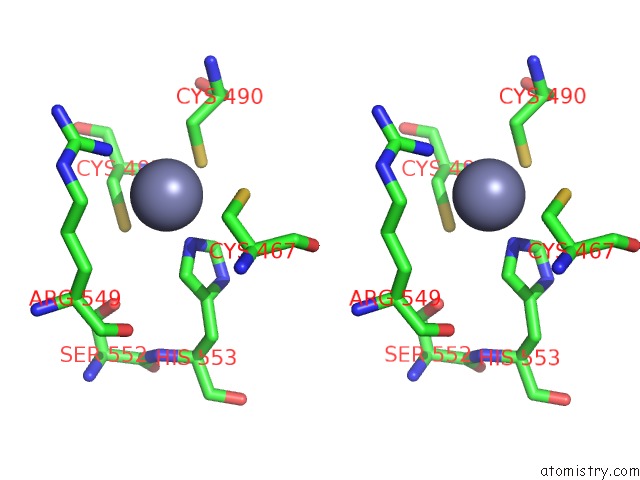
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of MEC1-DDC2 (Wild-Type) in Complex with Amp-Pnp within 5.0Å range:
|
Zinc binding site 2 out of 2 in 6z3a
Go back to
Zinc binding site 2 out
of 2 in the MEC1-DDC2 (Wild-Type) in Complex with Amp-Pnp
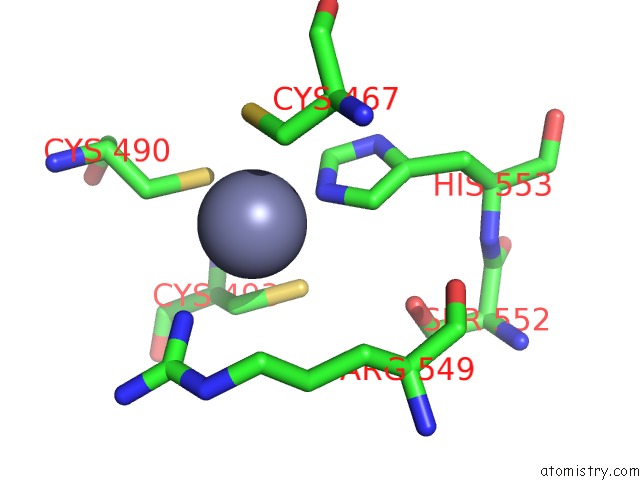
Mono view
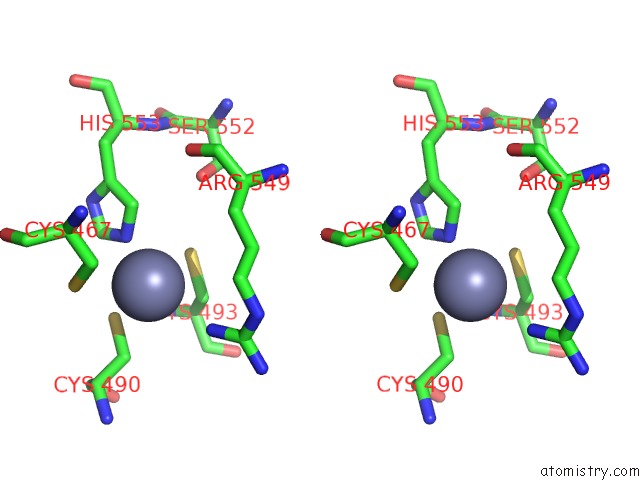
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 2 of MEC1-DDC2 (Wild-Type) in Complex with Amp-Pnp within 5.0Å range:
|
Reference:
E.A.Tannous,
L.A.Yates,
X.Zhang,
P.M.Burgers.
Mechanism of Auto-Inhibition and Activation of MEC1 Atr Checkpoint Kinase. Nat.Struct.Mol.Biol. 2020.
ISSN: ESSN 1545-9985
PubMed: 33169019
DOI: 10.1038/S41594-020-00522-0
Page generated: Thu Aug 21 21:38:53 2025
ISSN: ESSN 1545-9985
PubMed: 33169019
DOI: 10.1038/S41594-020-00522-0
Last articles
Zn in 7G70Zn in 7G6Z
Zn in 7G6X
Zn in 7G6Y
Zn in 7G6W
Zn in 7G6V
Zn in 7G6U
Zn in 7G6T
Zn in 7G6S
Zn in 7G6R