Zinc »
PDB 6wdv-6wq1 »
6wmp »
Zinc in PDB 6wmp: F. Tularensis RNAPS70-Igla Dna Complex
Enzymatic activity of F. Tularensis RNAPS70-Igla Dna Complex
All present enzymatic activity of F. Tularensis RNAPS70-Igla Dna Complex:
2.7.7.6;
2.7.7.6;
Other elements in 6wmp:
The structure of F. Tularensis RNAPS70-Igla Dna Complex also contains other interesting chemical elements:
| Magnesium | (Mg) | 1 atom |
Zinc Binding Sites:
The binding sites of Zinc atom in the F. Tularensis RNAPS70-Igla Dna Complex
(pdb code 6wmp). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total 2 binding sites of Zinc where determined in the F. Tularensis RNAPS70-Igla Dna Complex, PDB code: 6wmp:
Jump to Zinc binding site number: 1; 2;
In total 2 binding sites of Zinc where determined in the F. Tularensis RNAPS70-Igla Dna Complex, PDB code: 6wmp:
Jump to Zinc binding site number: 1; 2;
Zinc binding site 1 out of 2 in 6wmp
Go back to
Zinc binding site 1 out
of 2 in the F. Tularensis RNAPS70-Igla Dna Complex

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of F. Tularensis RNAPS70-Igla Dna Complex within 5.0Å range:
|
Zinc binding site 2 out of 2 in 6wmp
Go back to
Zinc binding site 2 out
of 2 in the F. Tularensis RNAPS70-Igla Dna Complex

Mono view
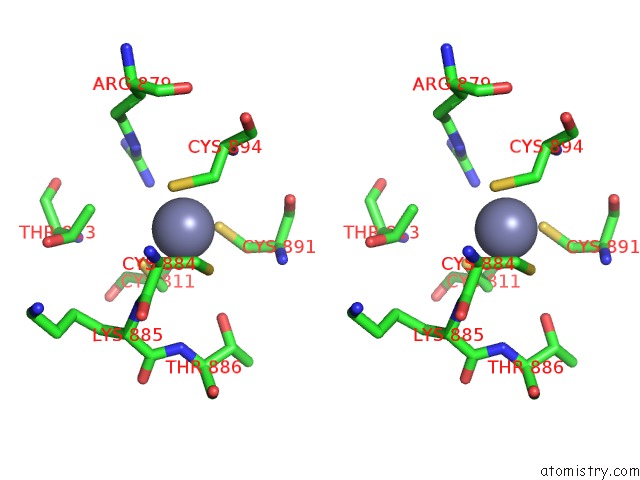
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 2 of F. Tularensis RNAPS70-Igla Dna Complex within 5.0Å range:
|
Reference:
B.A.Travis,
K.M.Ramsey,
S.M.Prezioso,
T.Tallo,
J.M.Wandzilak,
A.Hsu,
M.Borgnia,
A.Bartesaghi,
S.L.Dove,
R.G.Brennan,
M.A.Schumacher.
Structural Basis For Virulence Activation of Francisella Tularensis. Mol.Cell 2020.
ISSN: ISSN 1097-2765
PubMed: 33217319
DOI: 10.1016/J.MOLCEL.2020.10.035
Page generated: Tue Oct 29 10:13:43 2024
ISSN: ISSN 1097-2765
PubMed: 33217319
DOI: 10.1016/J.MOLCEL.2020.10.035
Last articles
Mg in 6B6WMg in 6B89
Mg in 6B8B
Mg in 6B6U
Mg in 6B66
Mg in 6B6H
Mg in 6B64
Mg in 6B63
Mg in 6B65
Mg in 6B62