Zinc »
PDB 6r57-6rdd »
6r7v »
Zinc in PDB 6r7v: Tannerella Forsythia Promirolysin Mutant E225A
Protein crystallography data
The structure of Tannerella Forsythia Promirolysin Mutant E225A, PDB code: 6r7v
was solved by
A.Rodriguez-Banqueri,
T.Guevara,
M.Ksiazek,
J.Potempa,
F.X.Gomis-Ruth,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 20.34 / 1.40 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 47.330, 67.250, 79.640, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 16.1 / 18.8 |
Other elements in 6r7v:
The structure of Tannerella Forsythia Promirolysin Mutant E225A also contains other interesting chemical elements:
| Calcium | (Ca) | 2 atoms |
Zinc Binding Sites:
The binding sites of Zinc atom in the Tannerella Forsythia Promirolysin Mutant E225A
(pdb code 6r7v). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total only one binding site of Zinc was determined in the Tannerella Forsythia Promirolysin Mutant E225A, PDB code: 6r7v:
In total only one binding site of Zinc was determined in the Tannerella Forsythia Promirolysin Mutant E225A, PDB code: 6r7v:
Zinc binding site 1 out of 1 in 6r7v
Go back to
Zinc binding site 1 out
of 1 in the Tannerella Forsythia Promirolysin Mutant E225A
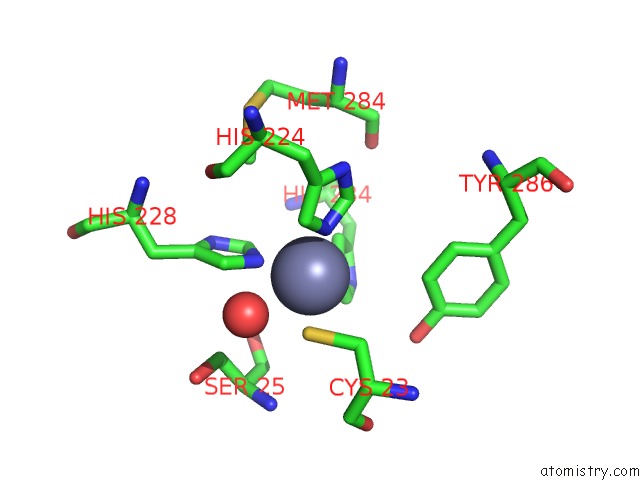
Mono view
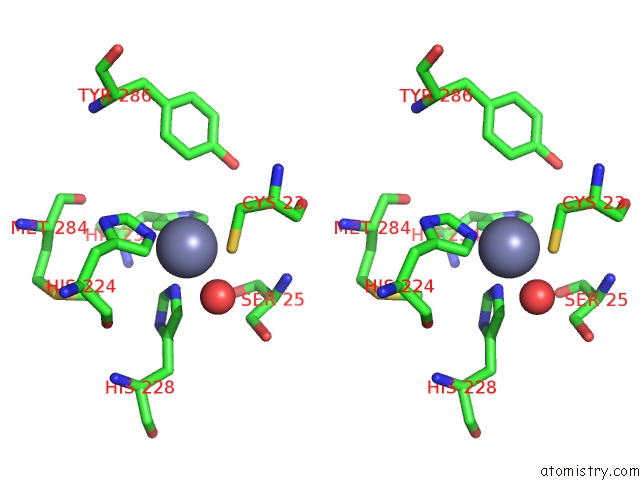
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Tannerella Forsythia Promirolysin Mutant E225A within 5.0Å range:
|
Reference:
T.Guevara,
A.Rodriguez-Banqueri,
M.Ksiazek,
J.Potempa,
F.X.Gomis-Ruth.
Structure-Based Mechanism of Cysteine-Switch Latency and of Catalysis By Pappalysin-Family Metallopeptidases Iucrj 2019.
ISSN: ESSN 2052-2525
Page generated: Thu Aug 21 19:03:03 2025
ISSN: ESSN 2052-2525
Last articles
Zn in 7AMVZn in 7AO9
Zn in 7AO8
Zn in 7AMS
Zn in 7AMR
Zn in 7AMQ
Zn in 7AMP
Zn in 7ALC
Zn in 7ALA
Zn in 7AKQ