Zinc »
PDB 6r55-6rda »
6r59 »
Zinc in PDB 6r59: Crystal Structure of Ppep-1(E143A/Y178F) in Complex with Substrate Peptide Ac-Evappvp-NH2
Enzymatic activity of Crystal Structure of Ppep-1(E143A/Y178F) in Complex with Substrate Peptide Ac-Evappvp-NH2
All present enzymatic activity of Crystal Structure of Ppep-1(E143A/Y178F) in Complex with Substrate Peptide Ac-Evappvp-NH2:
3.4.24.89;
3.4.24.89;
Protein crystallography data
The structure of Crystal Structure of Ppep-1(E143A/Y178F) in Complex with Substrate Peptide Ac-Evappvp-NH2, PDB code: 6r59
was solved by
C.Pichlo,
U.Baumann,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 40.49 / 1.65 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 37.110, 42.950, 122.240, 90.00, 96.60, 90.00 |
| R / Rfree (%) | 19.1 / 22 |
Zinc Binding Sites:
The binding sites of Zinc atom in the Crystal Structure of Ppep-1(E143A/Y178F) in Complex with Substrate Peptide Ac-Evappvp-NH2
(pdb code 6r59). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total 2 binding sites of Zinc where determined in the Crystal Structure of Ppep-1(E143A/Y178F) in Complex with Substrate Peptide Ac-Evappvp-NH2, PDB code: 6r59:
Jump to Zinc binding site number: 1; 2;
In total 2 binding sites of Zinc where determined in the Crystal Structure of Ppep-1(E143A/Y178F) in Complex with Substrate Peptide Ac-Evappvp-NH2, PDB code: 6r59:
Jump to Zinc binding site number: 1; 2;
Zinc binding site 1 out of 2 in 6r59
Go back to
Zinc binding site 1 out
of 2 in the Crystal Structure of Ppep-1(E143A/Y178F) in Complex with Substrate Peptide Ac-Evappvp-NH2
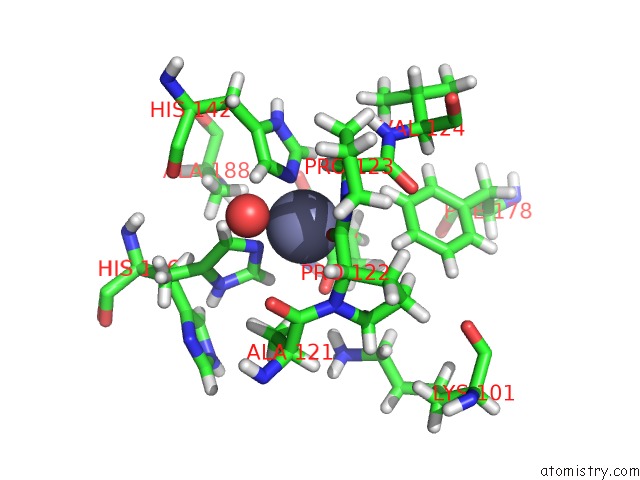
Mono view
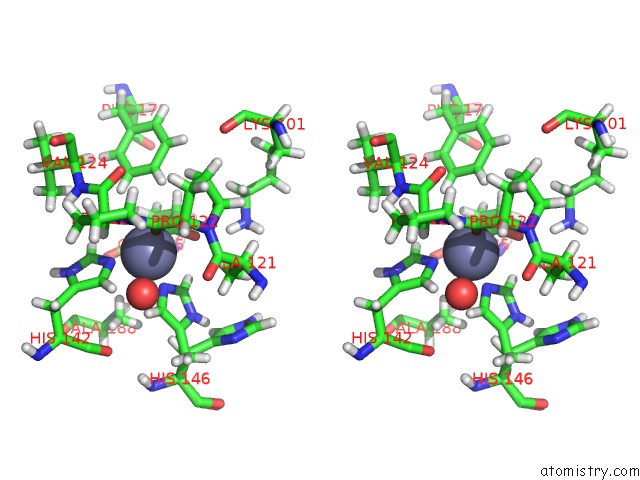
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Crystal Structure of Ppep-1(E143A/Y178F) in Complex with Substrate Peptide Ac-Evappvp-NH2 within 5.0Å range:
|
Zinc binding site 2 out of 2 in 6r59
Go back to
Zinc binding site 2 out
of 2 in the Crystal Structure of Ppep-1(E143A/Y178F) in Complex with Substrate Peptide Ac-Evappvp-NH2
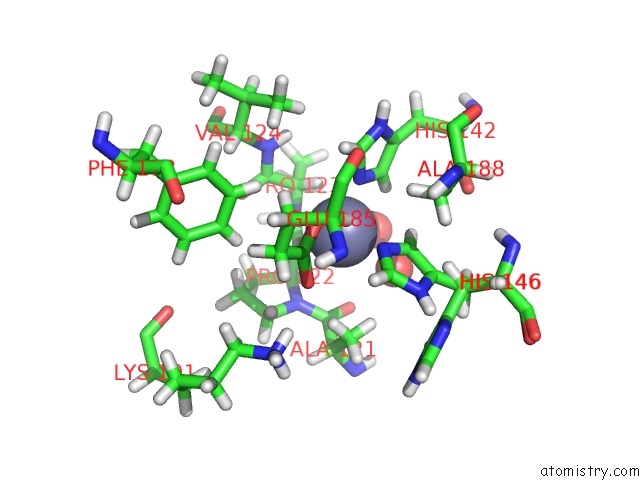
Mono view
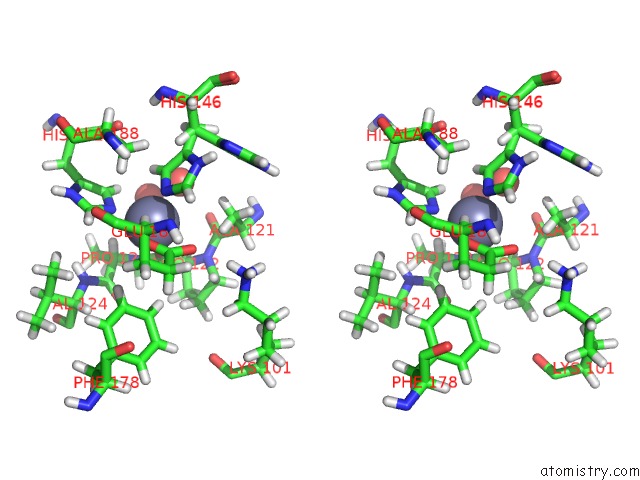
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 2 of Crystal Structure of Ppep-1(E143A/Y178F) in Complex with Substrate Peptide Ac-Evappvp-NH2 within 5.0Å range:
|
Reference:
C.Pichlo,
L.Juetten,
F.Wojtalla,
M.Schacherl,
D.Diaz,
U.Baumann.
Molecular Determinants of the Mechanism and Substrate Specificity Ofclostridium Difficileproline-Proline Endopeptidase-1. J.Biol.Chem. V. 294 11525 2019.
ISSN: ESSN 1083-351X
PubMed: 31182482
DOI: 10.1074/JBC.RA119.009029
Page generated: Tue Oct 29 06:01:55 2024
ISSN: ESSN 1083-351X
PubMed: 31182482
DOI: 10.1074/JBC.RA119.009029
Last articles
I in 4B9HI in 4AS2
I in 4AS5
I in 4AX2
I in 4ARR
I in 4AQ3
I in 4ARK
I in 4AP2
I in 4AIO
I in 4ANB