Zinc »
PDB 6p6g-6pi1 »
6pb5 »
Zinc in PDB 6pb5: The E. Coli Class-II Cap-Dependent Transcription Activation Complex at the State 1 Architecture
Enzymatic activity of The E. Coli Class-II Cap-Dependent Transcription Activation Complex at the State 1 Architecture
All present enzymatic activity of The E. Coli Class-II Cap-Dependent Transcription Activation Complex at the State 1 Architecture:
2.7.7.6;
2.7.7.6;
Other elements in 6pb5:
The structure of The E. Coli Class-II Cap-Dependent Transcription Activation Complex at the State 1 Architecture also contains other interesting chemical elements:
| Magnesium | (Mg) | 1 atom |
Zinc Binding Sites:
The binding sites of Zinc atom in the The E. Coli Class-II Cap-Dependent Transcription Activation Complex at the State 1 Architecture
(pdb code 6pb5). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total 2 binding sites of Zinc where determined in the The E. Coli Class-II Cap-Dependent Transcription Activation Complex at the State 1 Architecture, PDB code: 6pb5:
Jump to Zinc binding site number: 1; 2;
In total 2 binding sites of Zinc where determined in the The E. Coli Class-II Cap-Dependent Transcription Activation Complex at the State 1 Architecture, PDB code: 6pb5:
Jump to Zinc binding site number: 1; 2;
Zinc binding site 1 out of 2 in 6pb5
Go back to
Zinc binding site 1 out
of 2 in the The E. Coli Class-II Cap-Dependent Transcription Activation Complex at the State 1 Architecture
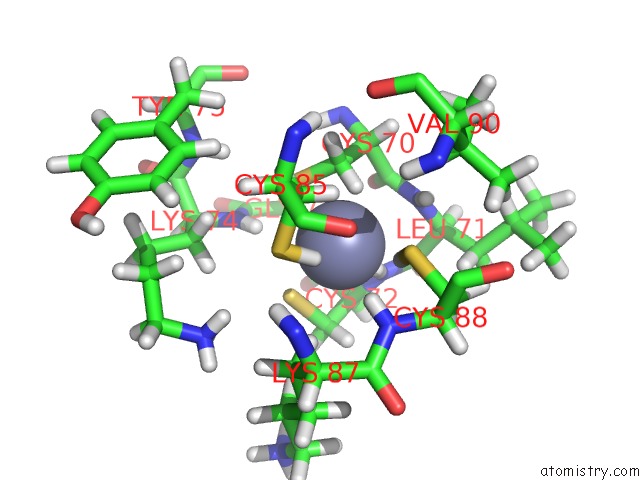
Mono view
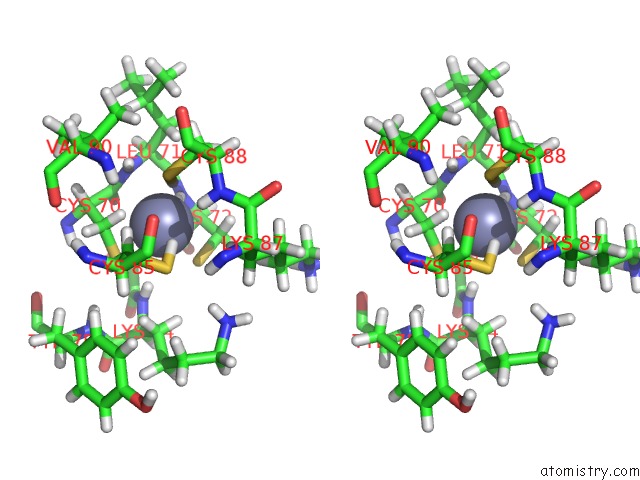
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of The E. Coli Class-II Cap-Dependent Transcription Activation Complex at the State 1 Architecture within 5.0Å range:
|
Zinc binding site 2 out of 2 in 6pb5
Go back to
Zinc binding site 2 out
of 2 in the The E. Coli Class-II Cap-Dependent Transcription Activation Complex at the State 1 Architecture
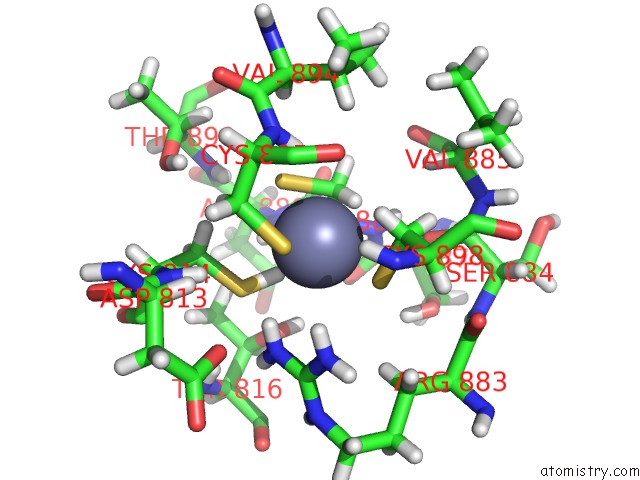
Mono view
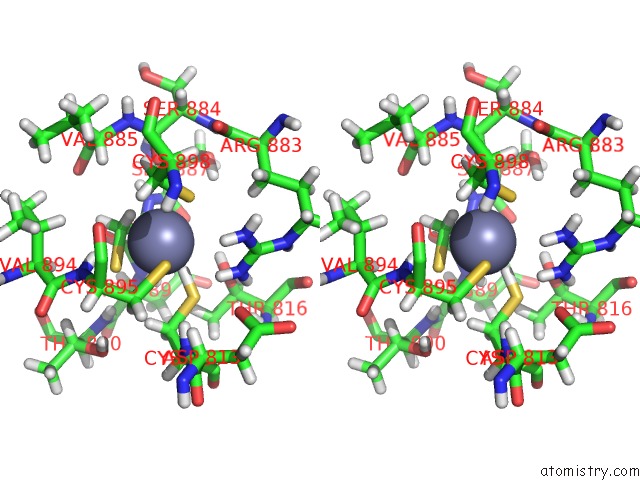
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 2 of The E. Coli Class-II Cap-Dependent Transcription Activation Complex at the State 1 Architecture within 5.0Å range:
|
Reference:
W.Shi,
Y.Jiang,
Y.Deng,
Z.Dong,
B.Liu.
Visualization of Two Architectures in Class-II Cap-Dependent Transcription Activation Plos Biol. 2020.
ISSN: ESSN 1545-7885
Page generated: Tue Oct 29 04:56:29 2024
ISSN: ESSN 1545-7885
Last articles
K in 3K2RK in 3K14
K in 3K13
K in 3JYC
K in 3JZO
K in 3JZP
K in 3ISE
K in 3IS7
K in 3IS8
K in 3JW4