Zinc »
PDB 6nl8-6o5g »
6nq3 »
Zinc in PDB 6nq3: Structure of A Chromatin Complex
Protein crystallography data
The structure of Structure of A Chromatin Complex, PDB code: 6nq3
was solved by
S.Chen,
L.Jiao,
X.Liu,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 46.60 / 2.89 |
| Space group | C 2 2 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 127.710, 139.600, 268.080, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 17 / 23 |
Zinc Binding Sites:
The binding sites of Zinc atom in the Structure of A Chromatin Complex
(pdb code 6nq3). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total 2 binding sites of Zinc where determined in the Structure of A Chromatin Complex, PDB code: 6nq3:
Jump to Zinc binding site number: 1; 2;
In total 2 binding sites of Zinc where determined in the Structure of A Chromatin Complex, PDB code: 6nq3:
Jump to Zinc binding site number: 1; 2;
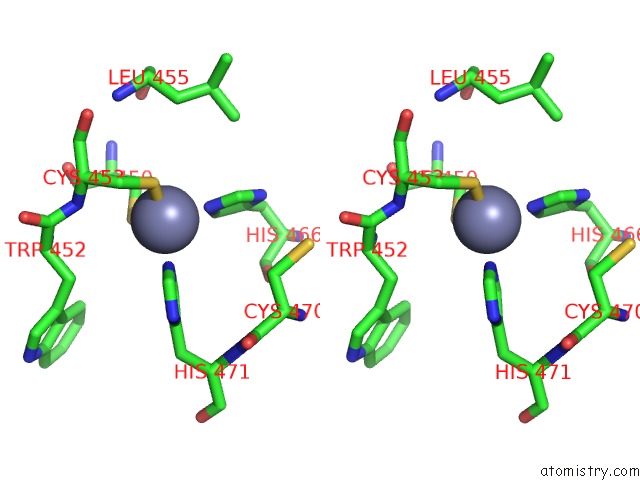
Zinc binding site 1 out of 2 in 6nq3
Go back to
Zinc binding site 1 out
of 2 in the Structure of A Chromatin Complex

Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Structure of A Chromatin Complex within 5.0Å range:
|
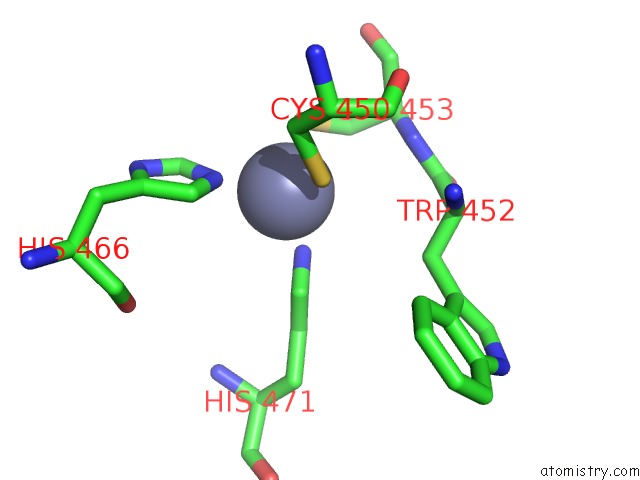
Zinc binding site 2 out of 2 in 6nq3
Go back to
Zinc binding site 2 out
of 2 in the Structure of A Chromatin Complex
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 2 of Structure of A Chromatin Complex within 5.0Å range:
|
Reference:
S.Chen,
L.Jiao,
X.Liu,
X.Yang,
X.Liu.
A Dimeric Structural Scaffold For PRC2-Pcl Targeting to Cpg Island Chromatin. Mol.Cell 2020.
ISSN: ISSN 1097-2765
PubMed: 31959557
DOI: 10.1016/J.MOLCEL.2019.12.019
Page generated: Tue Oct 29 03:58:11 2024
ISSN: ISSN 1097-2765
PubMed: 31959557
DOI: 10.1016/J.MOLCEL.2019.12.019
Last articles
I in 4RCFI in 4R11
I in 4RCE
I in 4RCD
I in 4R1T
I in 4QRP
I in 4QU3
I in 4PXS
I in 4QJQ
I in 4Q53