Zinc »
PDB 6jqx-6kal »
6k4y »
Zinc in PDB 6k4y: Cryoem Structure of Sigma Appropriation Complex
Enzymatic activity of Cryoem Structure of Sigma Appropriation Complex
All present enzymatic activity of Cryoem Structure of Sigma Appropriation Complex:
2.7.7.6;
2.7.7.6;
Other elements in 6k4y:
The structure of Cryoem Structure of Sigma Appropriation Complex also contains other interesting chemical elements:
| Magnesium | (Mg) | 1 atom |
Zinc Binding Sites:
The binding sites of Zinc atom in the Cryoem Structure of Sigma Appropriation Complex
(pdb code 6k4y). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total 2 binding sites of Zinc where determined in the Cryoem Structure of Sigma Appropriation Complex, PDB code: 6k4y:
Jump to Zinc binding site number: 1; 2;
In total 2 binding sites of Zinc where determined in the Cryoem Structure of Sigma Appropriation Complex, PDB code: 6k4y:
Jump to Zinc binding site number: 1; 2;
Zinc binding site 1 out of 2 in 6k4y
Go back to
Zinc binding site 1 out
of 2 in the Cryoem Structure of Sigma Appropriation Complex
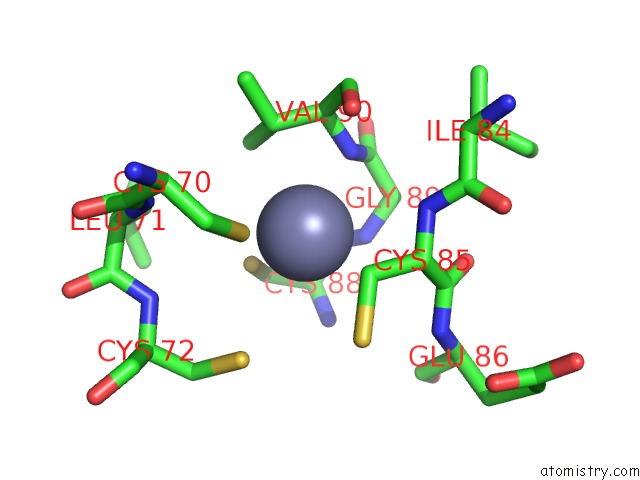
Mono view
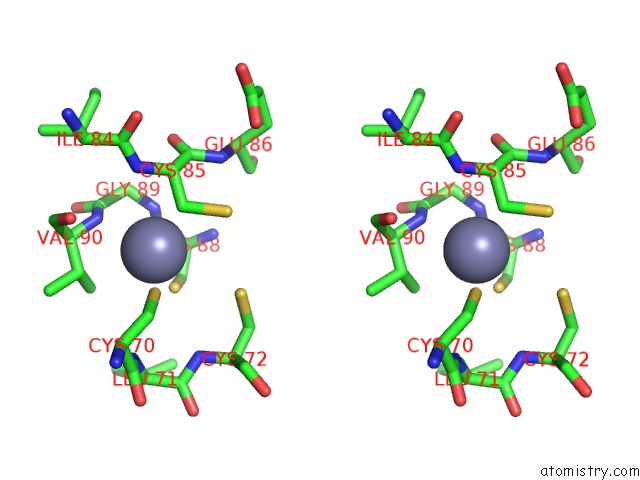
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Cryoem Structure of Sigma Appropriation Complex within 5.0Å range:
|
Zinc binding site 2 out of 2 in 6k4y
Go back to
Zinc binding site 2 out
of 2 in the Cryoem Structure of Sigma Appropriation Complex
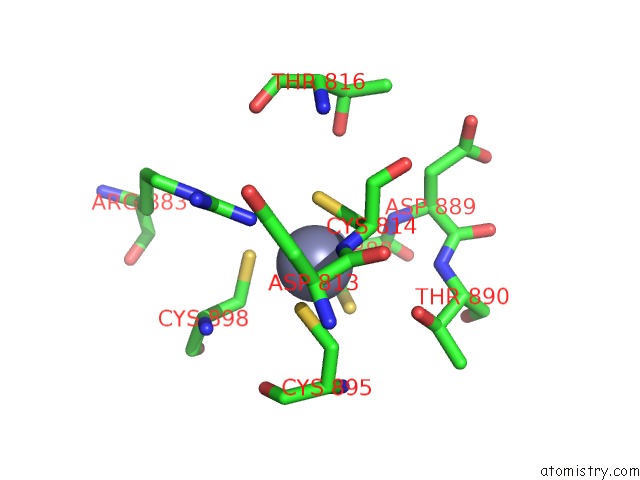
Mono view
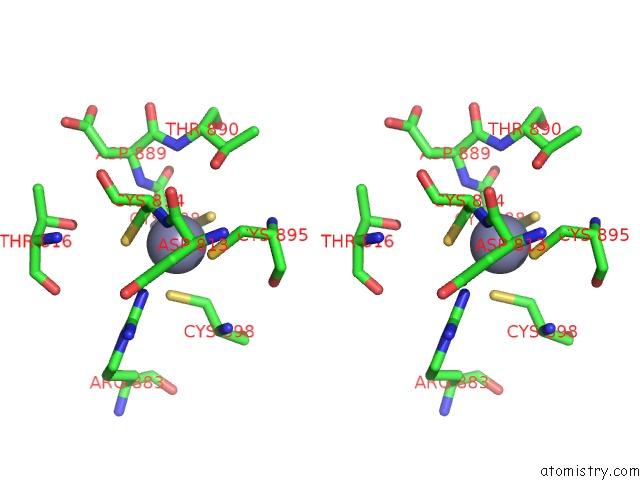
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 2 of Cryoem Structure of Sigma Appropriation Complex within 5.0Å range:
|
Reference:
J.Shi,
A.Wen,
M.Zhao,
L.You,
Y.Zhang,
Y.Feng.
Structural Basis of Sigma Appropriation. Nucleic Acids Res. V. 47 9423 2019.
ISSN: ESSN 1362-4962
PubMed: 31392983
DOI: 10.1093/NAR/GKZ682
Page generated: Tue Oct 29 01:32:15 2024
ISSN: ESSN 1362-4962
PubMed: 31392983
DOI: 10.1093/NAR/GKZ682
Last articles
Fe in 2YXOFe in 2YRS
Fe in 2YXC
Fe in 2YNM
Fe in 2YVJ
Fe in 2YP1
Fe in 2YU2
Fe in 2YU1
Fe in 2YQB
Fe in 2YOO