Zinc »
PDB 6a58-6ago »
6a5n »
Zinc in PDB 6a5n: Crystal Structure of Arabidopsis Thaliana SUVH6 in Complex with Methylated Dna
Enzymatic activity of Crystal Structure of Arabidopsis Thaliana SUVH6 in Complex with Methylated Dna
All present enzymatic activity of Crystal Structure of Arabidopsis Thaliana SUVH6 in Complex with Methylated Dna:
2.1.1.43;
2.1.1.43;
Protein crystallography data
The structure of Crystal Structure of Arabidopsis Thaliana SUVH6 in Complex with Methylated Dna, PDB code: 6a5n
was solved by
X.Li,
J.Du,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 33.59 / 2.40 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 74.137, 79.443, 107.845, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 23 / 27.9 |
Zinc Binding Sites:
The binding sites of Zinc atom in the Crystal Structure of Arabidopsis Thaliana SUVH6 in Complex with Methylated Dna
(pdb code 6a5n). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total 3 binding sites of Zinc where determined in the Crystal Structure of Arabidopsis Thaliana SUVH6 in Complex with Methylated Dna, PDB code: 6a5n:
Jump to Zinc binding site number: 1; 2; 3;
In total 3 binding sites of Zinc where determined in the Crystal Structure of Arabidopsis Thaliana SUVH6 in Complex with Methylated Dna, PDB code: 6a5n:
Jump to Zinc binding site number: 1; 2; 3;
Zinc binding site 1 out of 3 in 6a5n
Go back to
Zinc binding site 1 out
of 3 in the Crystal Structure of Arabidopsis Thaliana SUVH6 in Complex with Methylated Dna
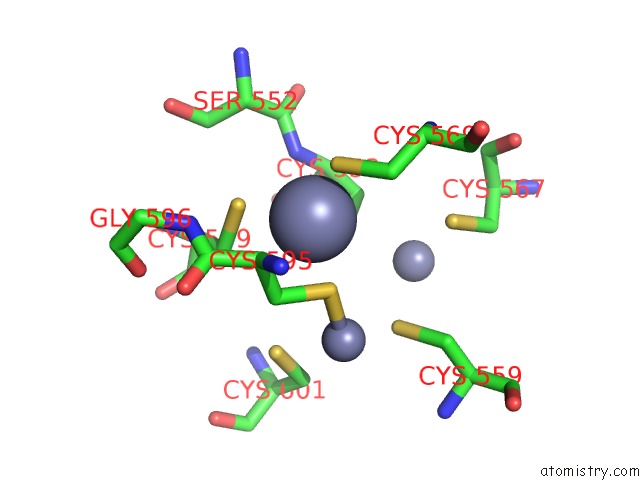
Mono view
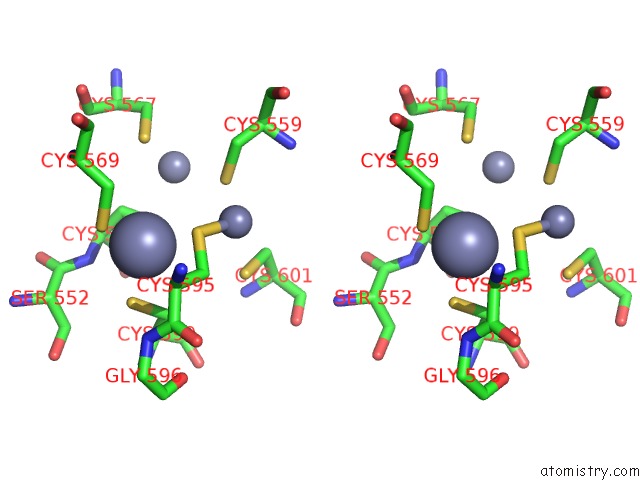
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Crystal Structure of Arabidopsis Thaliana SUVH6 in Complex with Methylated Dna within 5.0Å range:
|
Zinc binding site 2 out of 3 in 6a5n
Go back to
Zinc binding site 2 out
of 3 in the Crystal Structure of Arabidopsis Thaliana SUVH6 in Complex with Methylated Dna
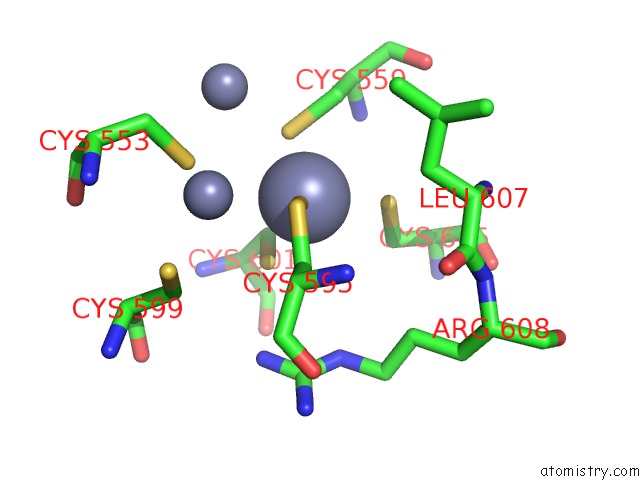
Mono view
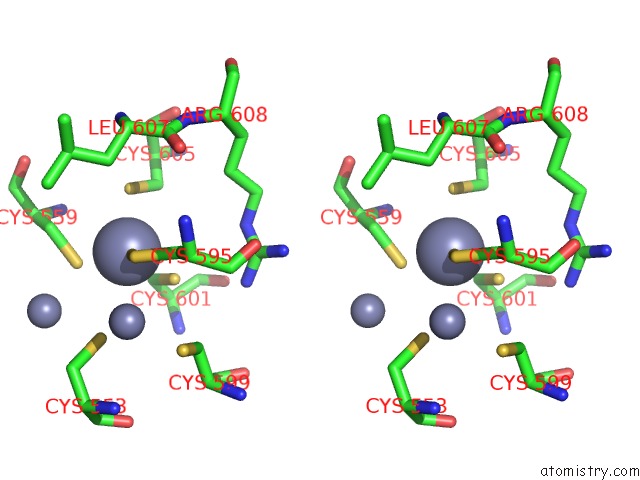
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 2 of Crystal Structure of Arabidopsis Thaliana SUVH6 in Complex with Methylated Dna within 5.0Å range:
|
Zinc binding site 3 out of 3 in 6a5n
Go back to
Zinc binding site 3 out
of 3 in the Crystal Structure of Arabidopsis Thaliana SUVH6 in Complex with Methylated Dna
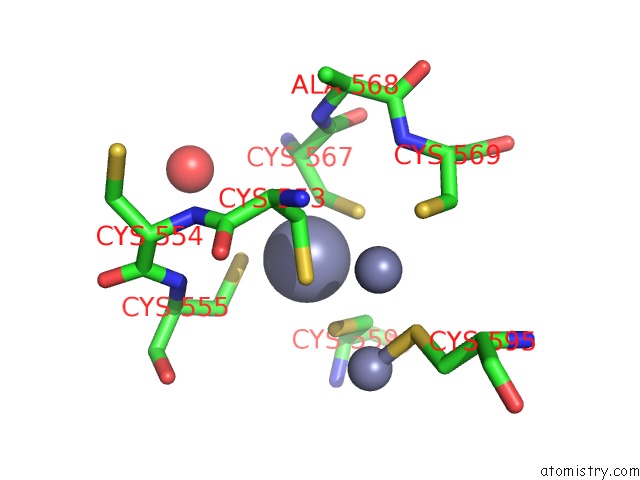
Mono view
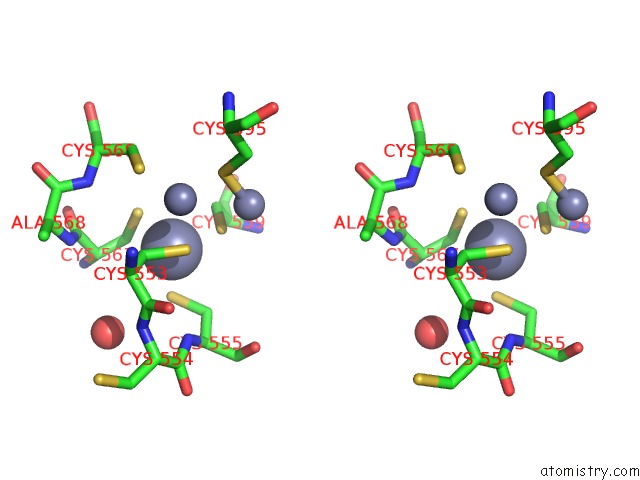
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 3 of Crystal Structure of Arabidopsis Thaliana SUVH6 in Complex with Methylated Dna within 5.0Å range:
|
Reference:
X.Li,
C.J.Harris,
Z.Zhong,
W.Chen,
R.Liu,
B.Jia,
Z.Wang,
S.Li,
S.E.Jacobsen,
J.Du.
Mechanistic Insights Into Plant Suvh Family H3K9 Methyltransferases and Their Binding to Context-Biased Non-Cg Dna Methylation. Proc. Natl. Acad. Sci. V. 115 E8793 2018U.S.A..
ISSN: ESSN 1091-6490
PubMed: 30150382
DOI: 10.1073/PNAS.1809841115
Page generated: Thu Aug 21 12:18:07 2025
ISSN: ESSN 1091-6490
PubMed: 30150382
DOI: 10.1073/PNAS.1809841115
Last articles
Zn in 6LI4Zn in 6LHN
Zn in 6LI2
Zn in 6LHD
Zn in 6LDG
Zn in 6LEH
Zn in 6LDF
Zn in 6LDI
Zn in 6LDE
Zn in 6LC1