Zinc »
PDB 5kb0-5kj1 »
5kdn »
Zinc in PDB 5kdn: Zmpb Metallopeptidase From Clostridium Perfringens
Protein crystallography data
The structure of Zmpb Metallopeptidase From Clostridium Perfringens, PDB code: 5kdn
was solved by
I.Noach,
E.Ficko-Blean,
C.Stuart,
A.B.Boraston,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 43.09 / 1.66 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 65.890, 68.020, 170.860, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 15.4 / 18 |
Zinc Binding Sites:
The binding sites of Zinc atom in the Zmpb Metallopeptidase From Clostridium Perfringens
(pdb code 5kdn). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total only one binding site of Zinc was determined in the Zmpb Metallopeptidase From Clostridium Perfringens, PDB code: 5kdn:
In total only one binding site of Zinc was determined in the Zmpb Metallopeptidase From Clostridium Perfringens, PDB code: 5kdn:
Zinc binding site 1 out of 1 in 5kdn
Go back to
Zinc binding site 1 out
of 1 in the Zmpb Metallopeptidase From Clostridium Perfringens

Mono view
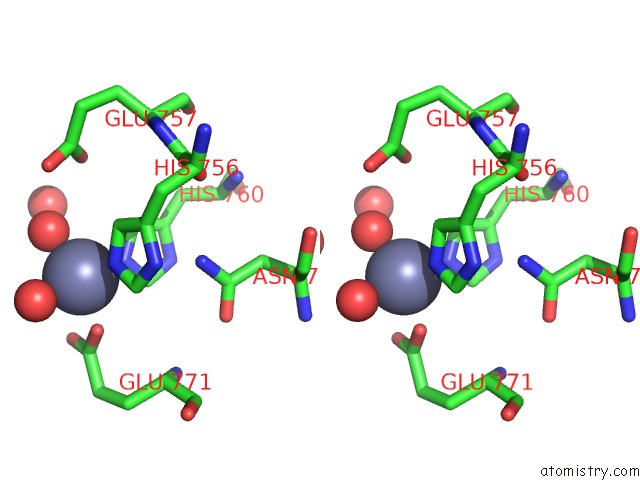
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Zmpb Metallopeptidase From Clostridium Perfringens within 5.0Å range:
|
Reference:
I.Noach,
E.Ficko-Blean,
B.Pluvinage,
C.Stuart,
M.L.Jenkins,
D.Brochu,
N.Buenbrazo,
W.Wakarchuk,
J.E.Burke,
M.Gilbert,
A.B.Boraston.
Recognition of Protein-Linked Glycans As A Determinant of Peptidase Activity. Proc. Natl. Acad. Sci. V. 114 E679 2017U.S.A..
ISSN: ESSN 1091-6490
PubMed: 28096352
DOI: 10.1073/PNAS.1615141114
Page generated: Sun Oct 27 20:18:34 2024
ISSN: ESSN 1091-6490
PubMed: 28096352
DOI: 10.1073/PNAS.1615141114
Last articles
Mg in 9BS6Mg in 9BTM
Mg in 9BR2
Mg in 9BQ6
Mg in 9BPM
Mg in 9BQ8
Mg in 9BPH
Mg in 9BP6
Mg in 9BPB
Mg in 9BPD