Zinc »
PDB 5jf2-5jmz »
5jg3 »
Zinc in PDB 5jg3: Human Carbonic Anhydrase II (121T/N67Q) Complexed with Benzo[D]Thiazole-2-Sulfonamide
Enzymatic activity of Human Carbonic Anhydrase II (121T/N67Q) Complexed with Benzo[D]Thiazole-2-Sulfonamide
All present enzymatic activity of Human Carbonic Anhydrase II (121T/N67Q) Complexed with Benzo[D]Thiazole-2-Sulfonamide:
4.2.1.1;
4.2.1.1;
Protein crystallography data
The structure of Human Carbonic Anhydrase II (121T/N67Q) Complexed with Benzo[D]Thiazole-2-Sulfonamide, PDB code: 5jg3
was solved by
J.M.Fox,
K.Kang,
M.Sastry,
W.Sherman,
B.Sankaran,
P.H.Zwart,
G.M.Whitesides,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 35.91 / 1.21 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 42.470, 41.690, 73.080, 90.00, 104.74, 90.00 |
| R / Rfree (%) | 13.2 / 15.2 |
Zinc Binding Sites:
The binding sites of Zinc atom in the Human Carbonic Anhydrase II (121T/N67Q) Complexed with Benzo[D]Thiazole-2-Sulfonamide
(pdb code 5jg3). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total only one binding site of Zinc was determined in the Human Carbonic Anhydrase II (121T/N67Q) Complexed with Benzo[D]Thiazole-2-Sulfonamide, PDB code: 5jg3:
In total only one binding site of Zinc was determined in the Human Carbonic Anhydrase II (121T/N67Q) Complexed with Benzo[D]Thiazole-2-Sulfonamide, PDB code: 5jg3:
Zinc binding site 1 out of 1 in 5jg3
Go back to
Zinc binding site 1 out
of 1 in the Human Carbonic Anhydrase II (121T/N67Q) Complexed with Benzo[D]Thiazole-2-Sulfonamide
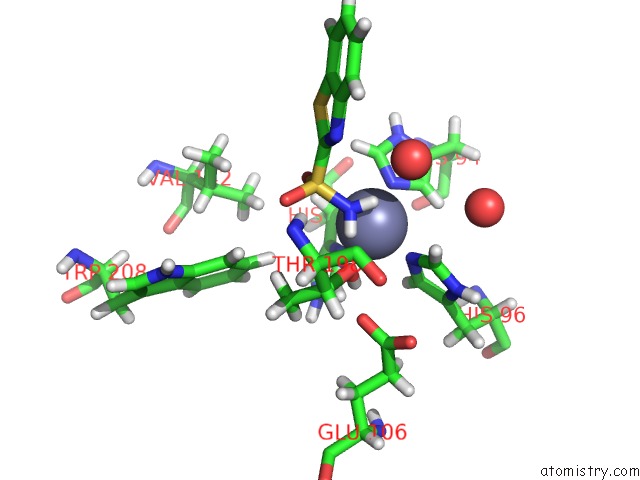
Mono view
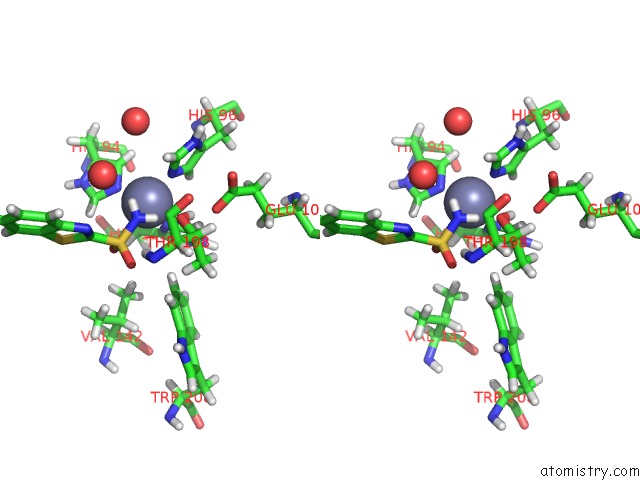
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Human Carbonic Anhydrase II (121T/N67Q) Complexed with Benzo[D]Thiazole-2-Sulfonamide within 5.0Å range:
|
Reference:
J.M.Fox,
K.Kang,
M.Sastry,
W.Sherman,
B.Sankaran,
P.H.Zwart,
G.M.Whitesides.
Water-Restructuring Mutations Can Reverse the Thermodynamic Signature of Ligand Binding to Human Carbonic Anhydrase. Angew. Chem. Int. Ed. Engl. V. 56 3833 2017.
ISSN: ESSN 1521-3773
PubMed: 28252841
DOI: 10.1002/ANIE.201609409
Page generated: Sun Oct 27 18:54:35 2024
ISSN: ESSN 1521-3773
PubMed: 28252841
DOI: 10.1002/ANIE.201609409
Last articles
Mg in 3G73Mg in 3G37
Mg in 3G6Y
Mg in 3G6X
Mg in 3G6W
Mg in 3G6V
Mg in 3G6K
Mg in 3G5A
Mg in 3G5S
Mg in 3G58