Zinc »
PDB 4l61-4leq »
4lal »
Zinc in PDB 4lal: Crystal Structure of Cordyceps Militaris Idcase D323A Mutant in Complex with 5-Carboxyl-Uracil
Protein crystallography data
The structure of Crystal Structure of Cordyceps Militaris Idcase D323A Mutant in Complex with 5-Carboxyl-Uracil, PDB code: 4lal
was solved by
S.Xu,
W.Li,
J.Zhu,
J.Ding,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 47.96 / 2.10 |
| Space group | C 1 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 192.266, 54.301, 143.776, 90.00, 121.04, 90.00 |
| R / Rfree (%) | 17.5 / 22 |
Zinc Binding Sites:
The binding sites of Zinc atom in the Crystal Structure of Cordyceps Militaris Idcase D323A Mutant in Complex with 5-Carboxyl-Uracil
(pdb code 4lal). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total 4 binding sites of Zinc where determined in the Crystal Structure of Cordyceps Militaris Idcase D323A Mutant in Complex with 5-Carboxyl-Uracil, PDB code: 4lal:
Jump to Zinc binding site number: 1; 2; 3; 4;
In total 4 binding sites of Zinc where determined in the Crystal Structure of Cordyceps Militaris Idcase D323A Mutant in Complex with 5-Carboxyl-Uracil, PDB code: 4lal:
Jump to Zinc binding site number: 1; 2; 3; 4;
Zinc binding site 1 out of 4 in 4lal
Go back to
Zinc binding site 1 out
of 4 in the Crystal Structure of Cordyceps Militaris Idcase D323A Mutant in Complex with 5-Carboxyl-Uracil
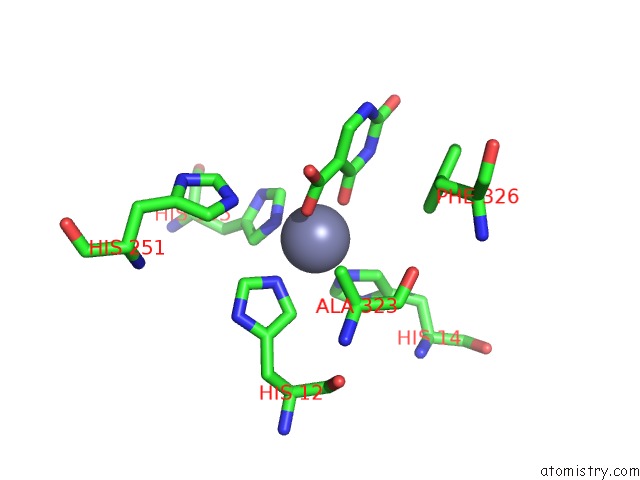
Mono view
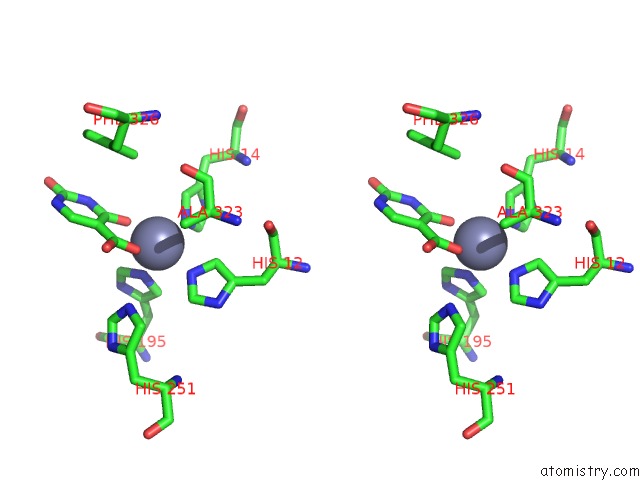
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Crystal Structure of Cordyceps Militaris Idcase D323A Mutant in Complex with 5-Carboxyl-Uracil within 5.0Å range:
|
Zinc binding site 2 out of 4 in 4lal
Go back to
Zinc binding site 2 out
of 4 in the Crystal Structure of Cordyceps Militaris Idcase D323A Mutant in Complex with 5-Carboxyl-Uracil
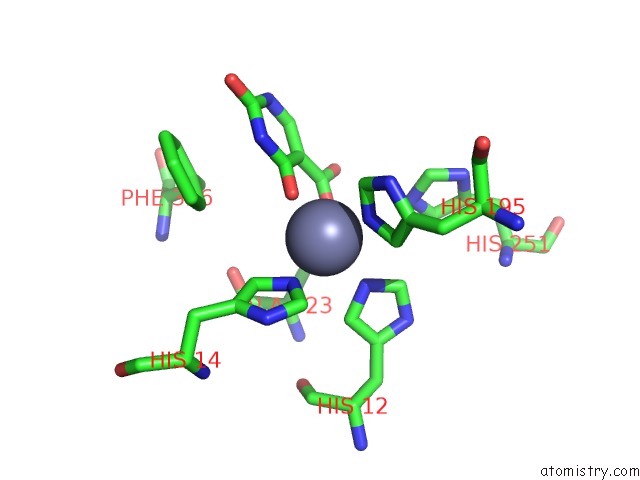
Mono view
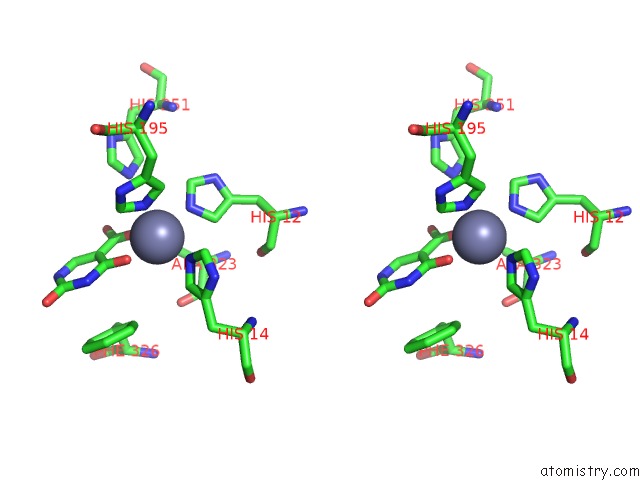
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 2 of Crystal Structure of Cordyceps Militaris Idcase D323A Mutant in Complex with 5-Carboxyl-Uracil within 5.0Å range:
|
Zinc binding site 3 out of 4 in 4lal
Go back to
Zinc binding site 3 out
of 4 in the Crystal Structure of Cordyceps Militaris Idcase D323A Mutant in Complex with 5-Carboxyl-Uracil
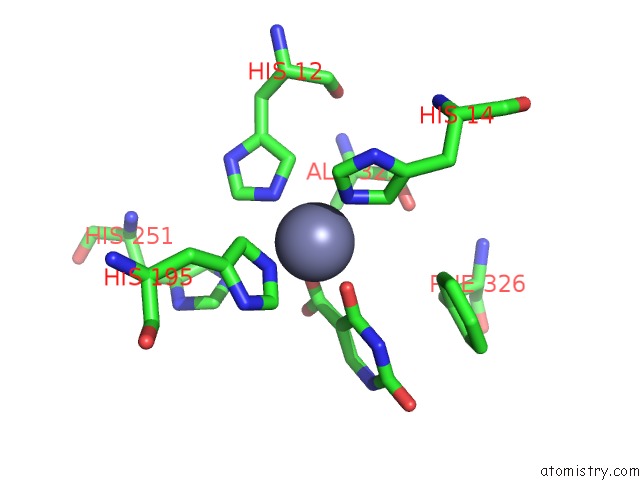
Mono view
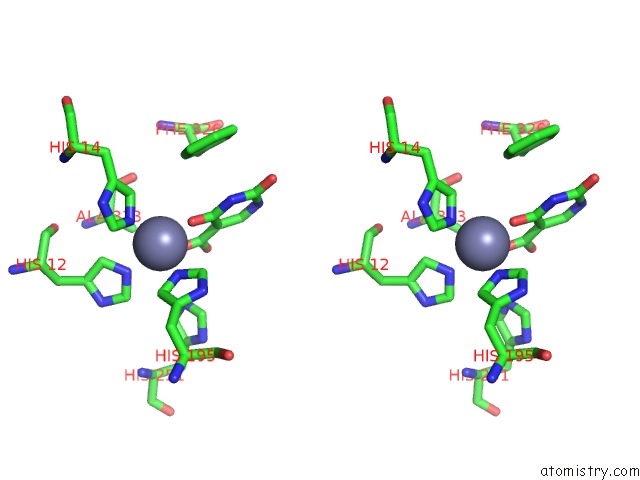
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 3 of Crystal Structure of Cordyceps Militaris Idcase D323A Mutant in Complex with 5-Carboxyl-Uracil within 5.0Å range:
|
Zinc binding site 4 out of 4 in 4lal
Go back to
Zinc binding site 4 out
of 4 in the Crystal Structure of Cordyceps Militaris Idcase D323A Mutant in Complex with 5-Carboxyl-Uracil
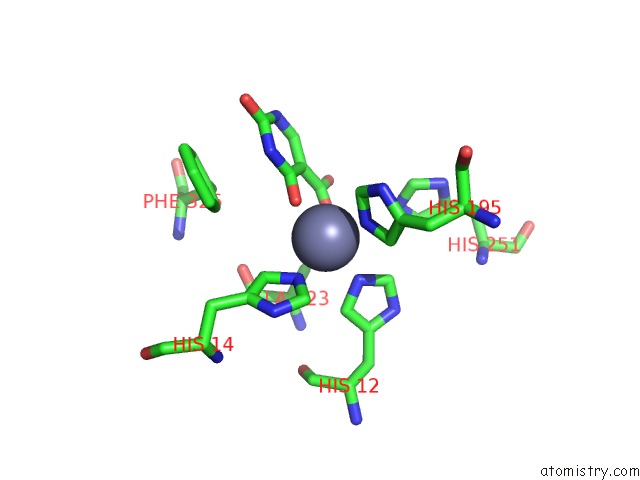
Mono view
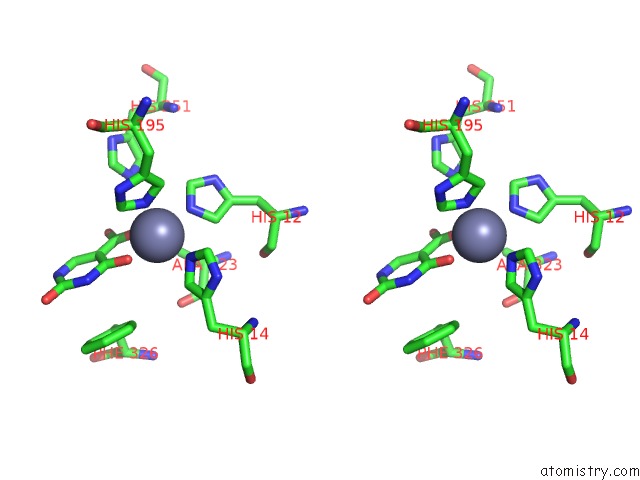
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 4 of Crystal Structure of Cordyceps Militaris Idcase D323A Mutant in Complex with 5-Carboxyl-Uracil within 5.0Å range:
|
Reference:
S.Xu,
W.Li,
J.Zhu,
R.Wang,
Z.Li,
G.L.Xu,
J.Ding.
Crystal Structures of Isoorotate Decarboxylases Reveal A Novel Catalytic Mechanism of 5-Carboxyl-Uracil Decarboxylation and Shed Light on the Search For Dna Decarboxylase. Cell Res. V. 23 1296 2013.
ISSN: ISSN 1001-0602
PubMed: 23917530
DOI: 10.1038/CR.2013.107
Page generated: Sun Oct 27 01:39:10 2024
ISSN: ISSN 1001-0602
PubMed: 23917530
DOI: 10.1038/CR.2013.107
Last articles
Mg in 7O0QMg in 7O04
Mg in 7NZ0
Mg in 7NYZ
Mg in 7NYX
Mg in 7NYW
Mg in 7NY1
Mg in 7NYG
Mg in 7NYF
Mg in 7NYA