Zinc »
PDB 3tge-3tus »
3tn2 »
Zinc in PDB 3tn2: Structure Analysis of MIP1-Beta P8A
Protein crystallography data
The structure of Structure Analysis of MIP1-Beta P8A, PDB code: 3tn2
was solved by
Q.Guo,
W.J.Tang,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 29.45 / 1.60 |
| Space group | C 1 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 51.062, 36.972, 31.061, 90.00, 108.54, 90.00 |
| R / Rfree (%) | 15.7 / 19.9 |
Zinc Binding Sites:
The binding sites of Zinc atom in the Structure Analysis of MIP1-Beta P8A
(pdb code 3tn2). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total 3 binding sites of Zinc where determined in the Structure Analysis of MIP1-Beta P8A, PDB code: 3tn2:
Jump to Zinc binding site number: 1; 2; 3;
In total 3 binding sites of Zinc where determined in the Structure Analysis of MIP1-Beta P8A, PDB code: 3tn2:
Jump to Zinc binding site number: 1; 2; 3;
Zinc binding site 1 out of 3 in 3tn2
Go back to
Zinc binding site 1 out
of 3 in the Structure Analysis of MIP1-Beta P8A
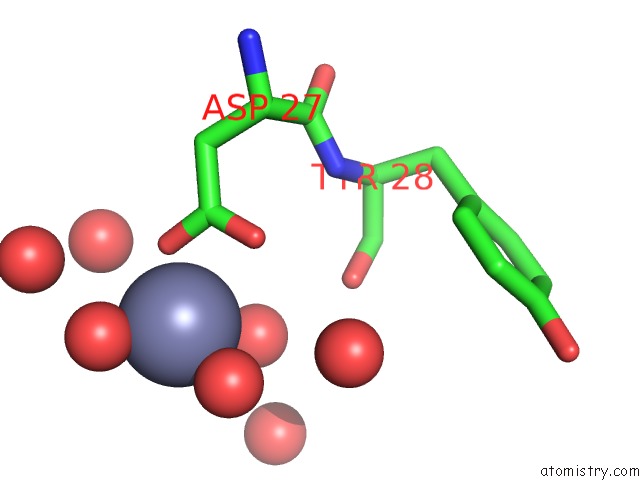
Mono view
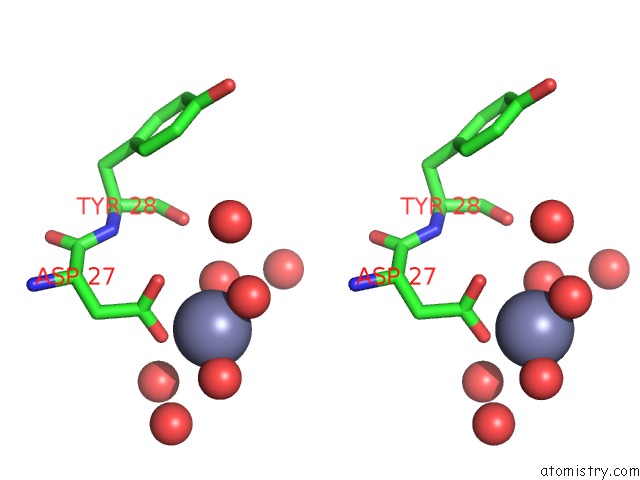
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Structure Analysis of MIP1-Beta P8A within 5.0Å range:
|
Zinc binding site 2 out of 3 in 3tn2
Go back to
Zinc binding site 2 out
of 3 in the Structure Analysis of MIP1-Beta P8A
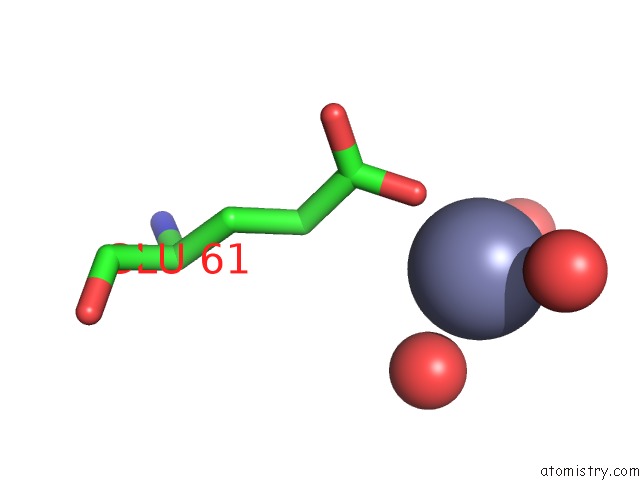
Mono view
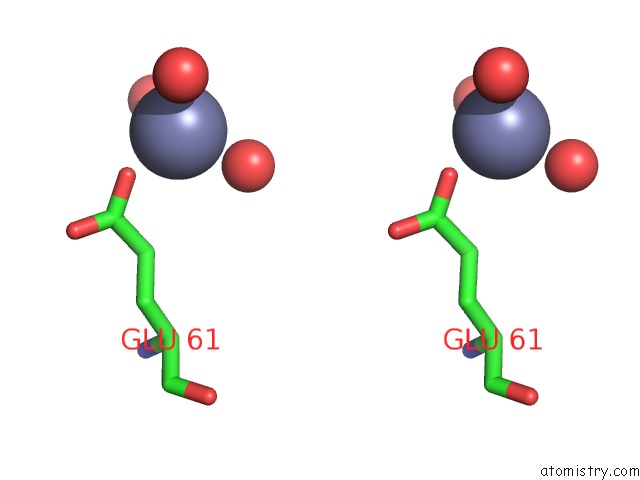
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 2 of Structure Analysis of MIP1-Beta P8A within 5.0Å range:
|
Zinc binding site 3 out of 3 in 3tn2
Go back to
Zinc binding site 3 out
of 3 in the Structure Analysis of MIP1-Beta P8A
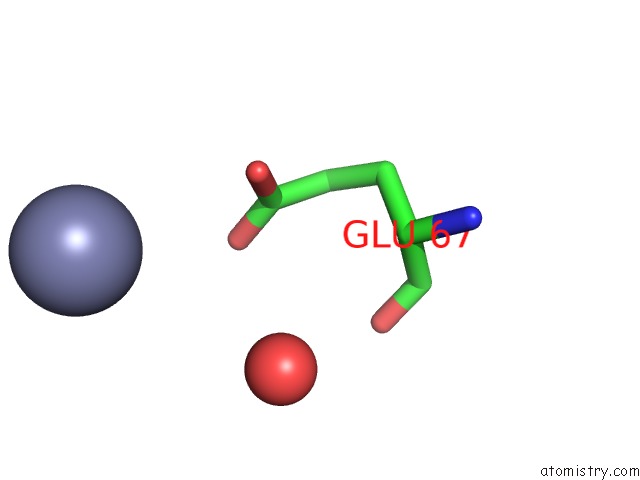
Mono view
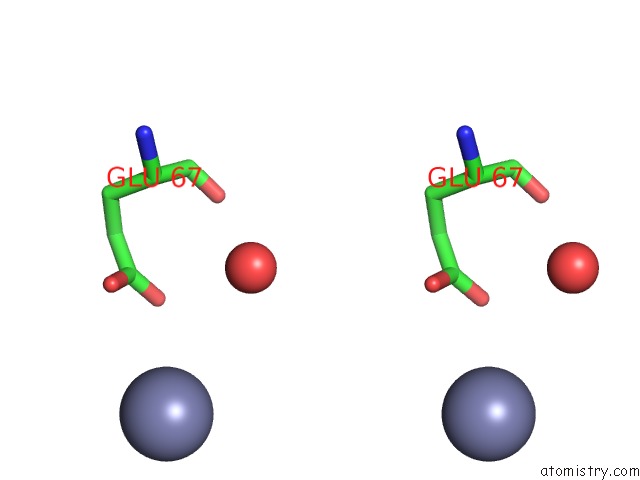
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 3 of Structure Analysis of MIP1-Beta P8A within 5.0Å range:
|
Reference:
W.G.Liang,
M.Ren,
F.Zhao,
W.J.Tang.
Structures of Human CCL18, CCL3, and CCL4 Reveal Molecular Determinants For Quaternary Structures and Sensitivity to Insulin-Degrading Enzyme. J.Mol.Biol. V. 427 1345 2015.
ISSN: ISSN 0022-2836
PubMed: 25636406
DOI: 10.1016/J.JMB.2015.01.012
Page generated: Wed Aug 20 14:30:46 2025
ISSN: ISSN 0022-2836
PubMed: 25636406
DOI: 10.1016/J.JMB.2015.01.012
Last articles
Zn in 4KCHZn in 4KAY
Zn in 4KBP
Zn in 4KAV
Zn in 4KB6
Zn in 4KA9
Zn in 4KA7
Zn in 4KAP
Zn in 4KA8
Zn in 4K95