Zinc »
PDB 3tge-3tus »
3tio »
Zinc in PDB 3tio: Crystal Structures of Yrda From Escherichia Coli, A Homologous Protein of Gamma-Class Carbonic Anhydrase, Show Possible Allosteric Conformations
Protein crystallography data
The structure of Crystal Structures of Yrda From Escherichia Coli, A Homologous Protein of Gamma-Class Carbonic Anhydrase, Show Possible Allosteric Conformations, PDB code: 3tio
was solved by
H.M.Park,
J.W.Choi,
J.E.Lee,
C.H.Jung,
B.Y.Kim,
J.S.Kim,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 50.00 / 1.41 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 55.976, 84.384, 97.758, 90.00, 93.18, 90.00 |
| R / Rfree (%) | 19.3 / 21 |
Zinc Binding Sites:
The binding sites of Zinc atom in the Crystal Structures of Yrda From Escherichia Coli, A Homologous Protein of Gamma-Class Carbonic Anhydrase, Show Possible Allosteric Conformations
(pdb code 3tio). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total 6 binding sites of Zinc where determined in the Crystal Structures of Yrda From Escherichia Coli, A Homologous Protein of Gamma-Class Carbonic Anhydrase, Show Possible Allosteric Conformations, PDB code: 3tio:
Jump to Zinc binding site number: 1; 2; 3; 4; 5; 6;
In total 6 binding sites of Zinc where determined in the Crystal Structures of Yrda From Escherichia Coli, A Homologous Protein of Gamma-Class Carbonic Anhydrase, Show Possible Allosteric Conformations, PDB code: 3tio:
Jump to Zinc binding site number: 1; 2; 3; 4; 5; 6;
Zinc binding site 1 out of 6 in 3tio
Go back to
Zinc binding site 1 out
of 6 in the Crystal Structures of Yrda From Escherichia Coli, A Homologous Protein of Gamma-Class Carbonic Anhydrase, Show Possible Allosteric Conformations
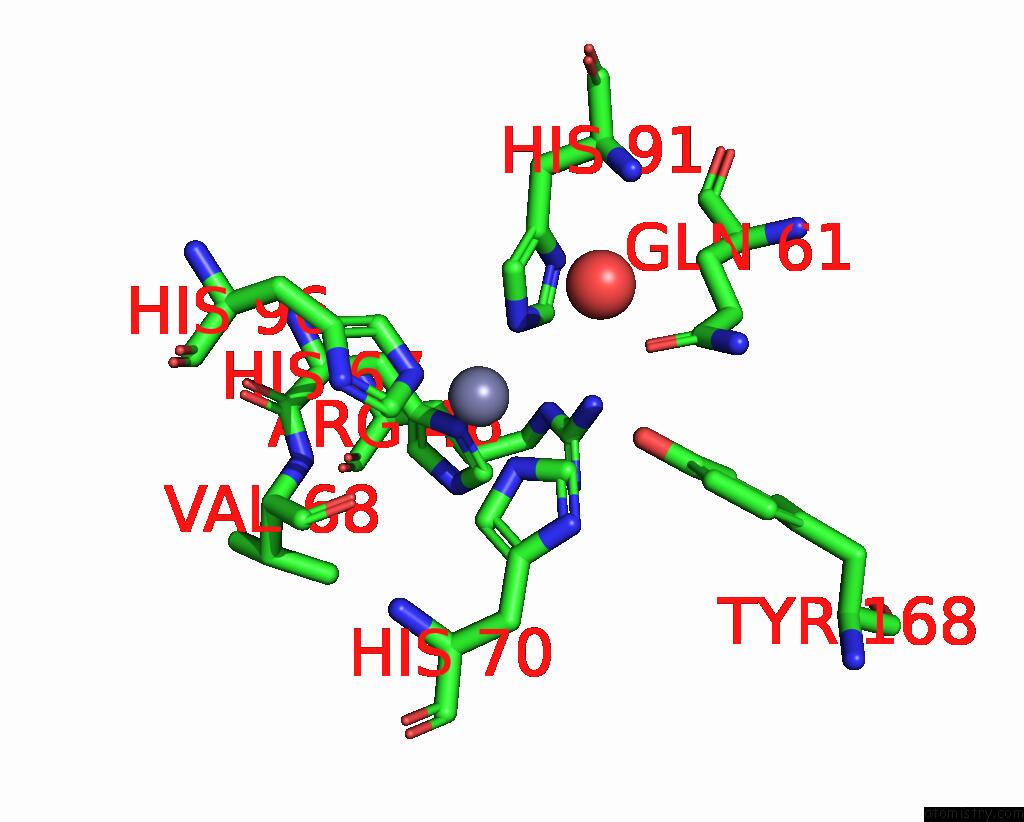
Mono view
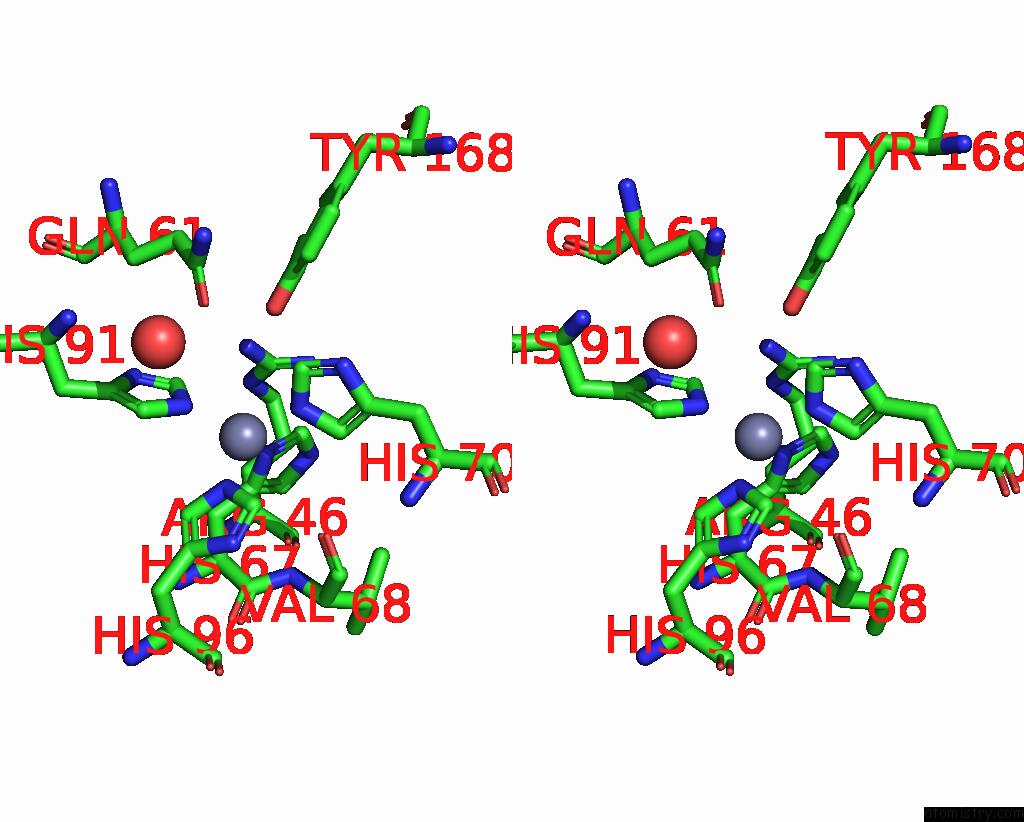
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Crystal Structures of Yrda From Escherichia Coli, A Homologous Protein of Gamma-Class Carbonic Anhydrase, Show Possible Allosteric Conformations within 5.0Å range:
|
Zinc binding site 2 out of 6 in 3tio
Go back to
Zinc binding site 2 out
of 6 in the Crystal Structures of Yrda From Escherichia Coli, A Homologous Protein of Gamma-Class Carbonic Anhydrase, Show Possible Allosteric Conformations
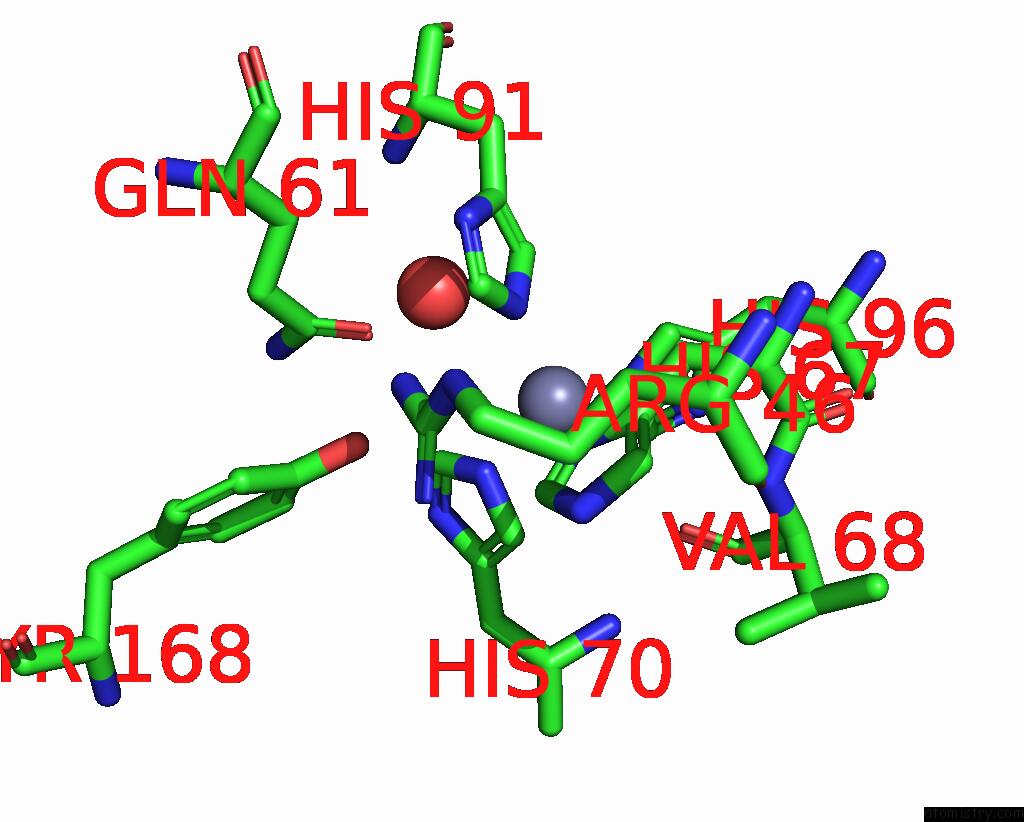
Mono view
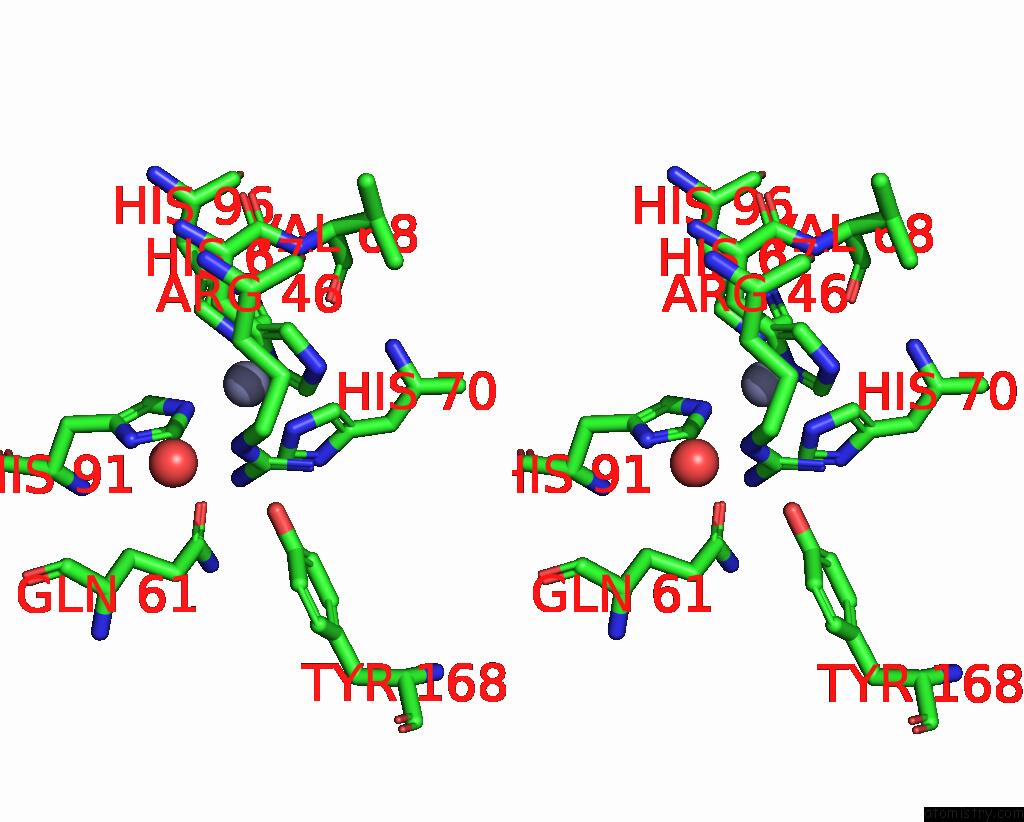
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 2 of Crystal Structures of Yrda From Escherichia Coli, A Homologous Protein of Gamma-Class Carbonic Anhydrase, Show Possible Allosteric Conformations within 5.0Å range:
|
Zinc binding site 3 out of 6 in 3tio
Go back to
Zinc binding site 3 out
of 6 in the Crystal Structures of Yrda From Escherichia Coli, A Homologous Protein of Gamma-Class Carbonic Anhydrase, Show Possible Allosteric Conformations
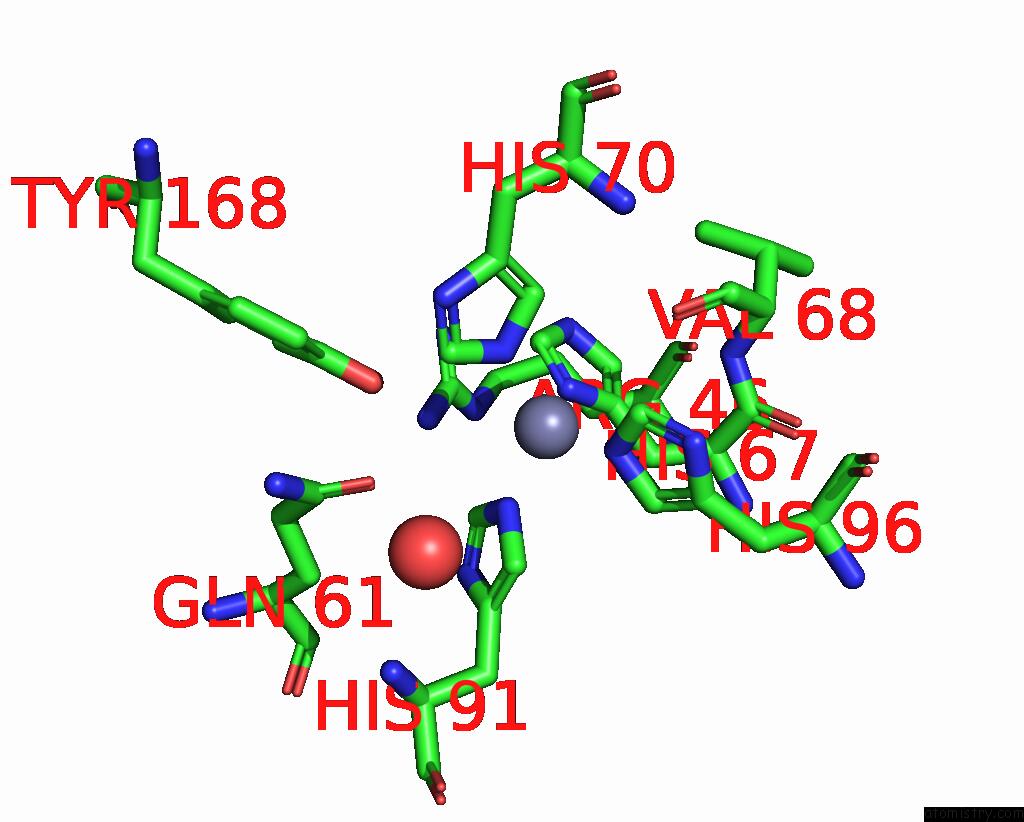
Mono view
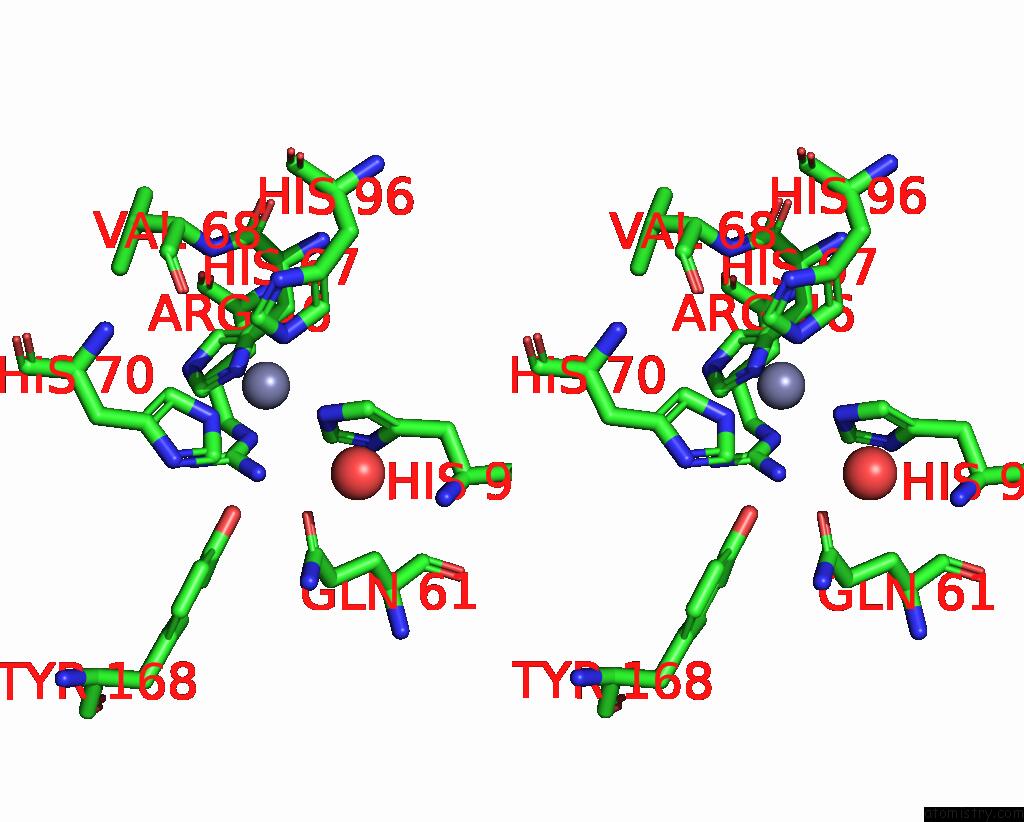
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 3 of Crystal Structures of Yrda From Escherichia Coli, A Homologous Protein of Gamma-Class Carbonic Anhydrase, Show Possible Allosteric Conformations within 5.0Å range:
|
Zinc binding site 4 out of 6 in 3tio
Go back to
Zinc binding site 4 out
of 6 in the Crystal Structures of Yrda From Escherichia Coli, A Homologous Protein of Gamma-Class Carbonic Anhydrase, Show Possible Allosteric Conformations
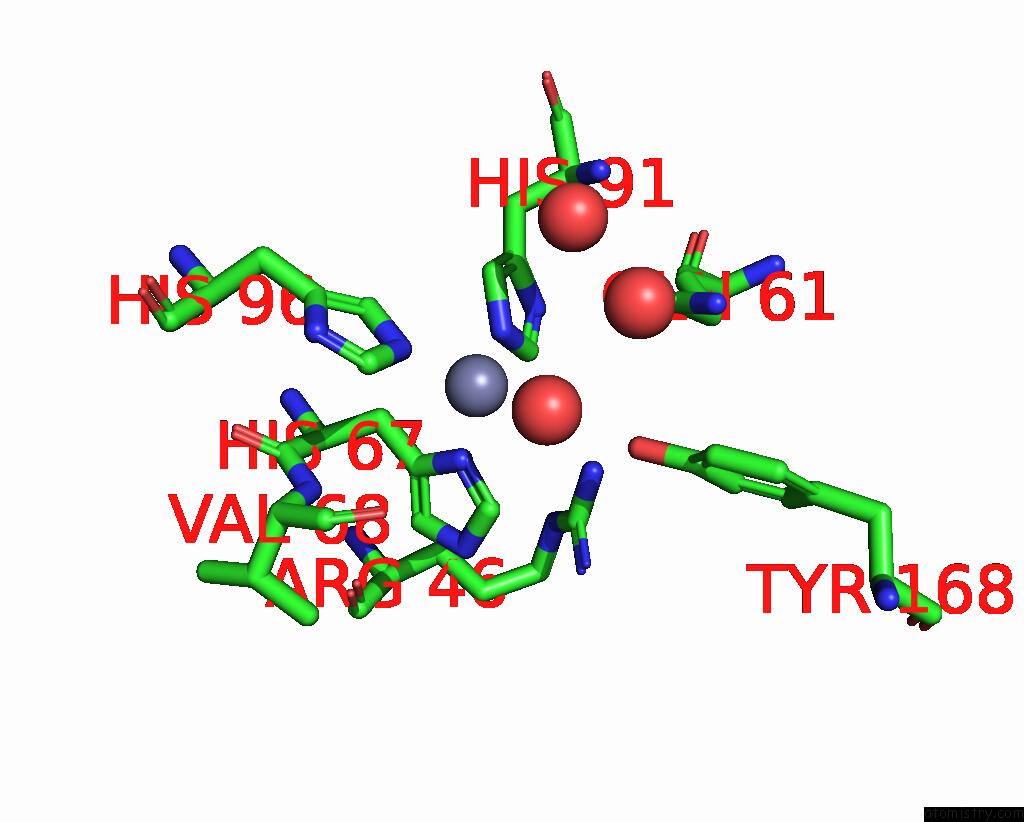
Mono view
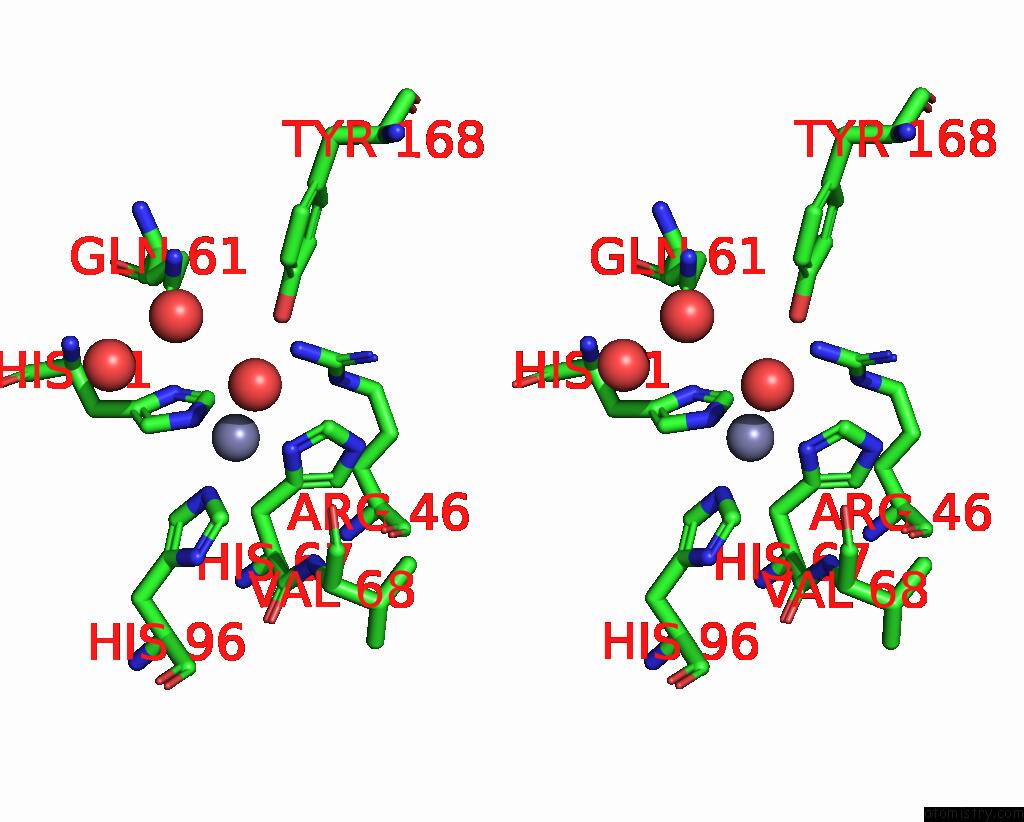
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 4 of Crystal Structures of Yrda From Escherichia Coli, A Homologous Protein of Gamma-Class Carbonic Anhydrase, Show Possible Allosteric Conformations within 5.0Å range:
|
Zinc binding site 5 out of 6 in 3tio
Go back to
Zinc binding site 5 out
of 6 in the Crystal Structures of Yrda From Escherichia Coli, A Homologous Protein of Gamma-Class Carbonic Anhydrase, Show Possible Allosteric Conformations
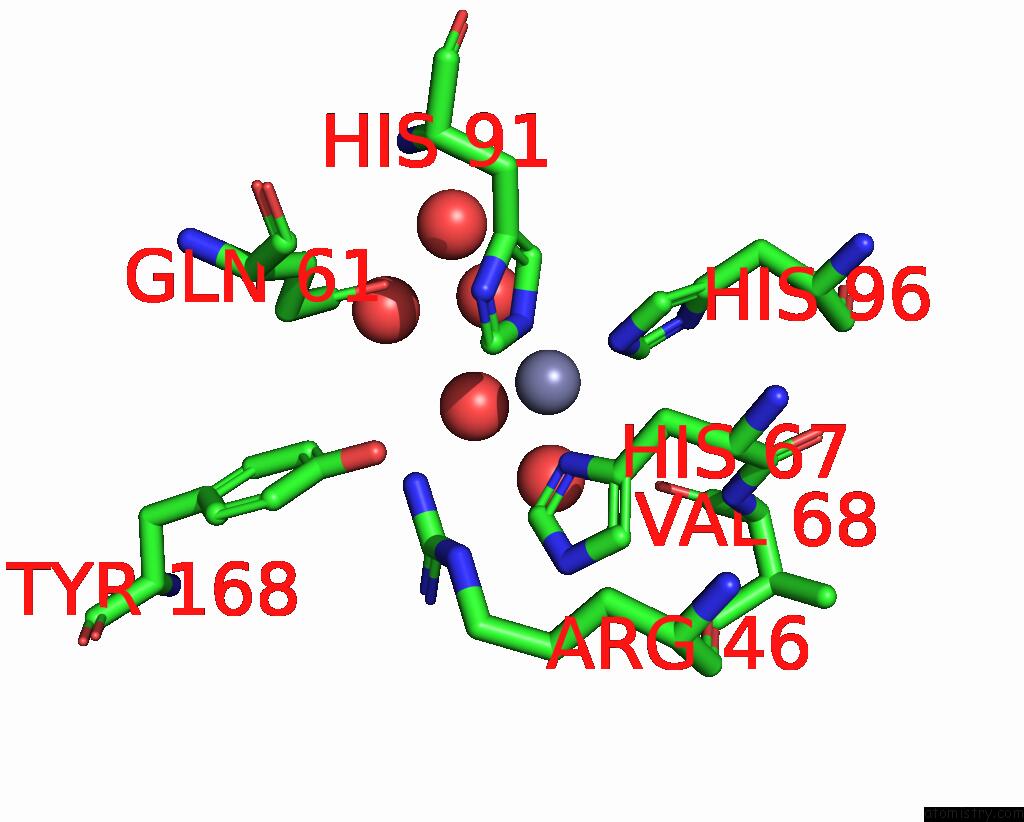
Mono view
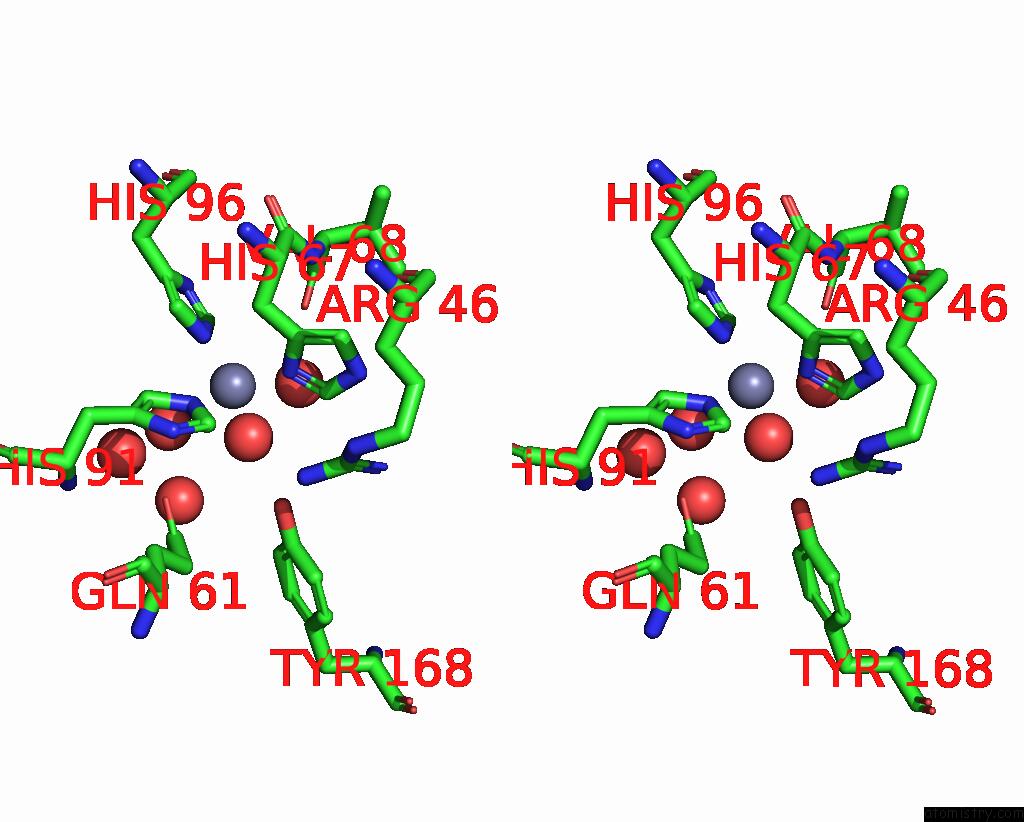
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 5 of Crystal Structures of Yrda From Escherichia Coli, A Homologous Protein of Gamma-Class Carbonic Anhydrase, Show Possible Allosteric Conformations within 5.0Å range:
|
Zinc binding site 6 out of 6 in 3tio
Go back to
Zinc binding site 6 out
of 6 in the Crystal Structures of Yrda From Escherichia Coli, A Homologous Protein of Gamma-Class Carbonic Anhydrase, Show Possible Allosteric Conformations
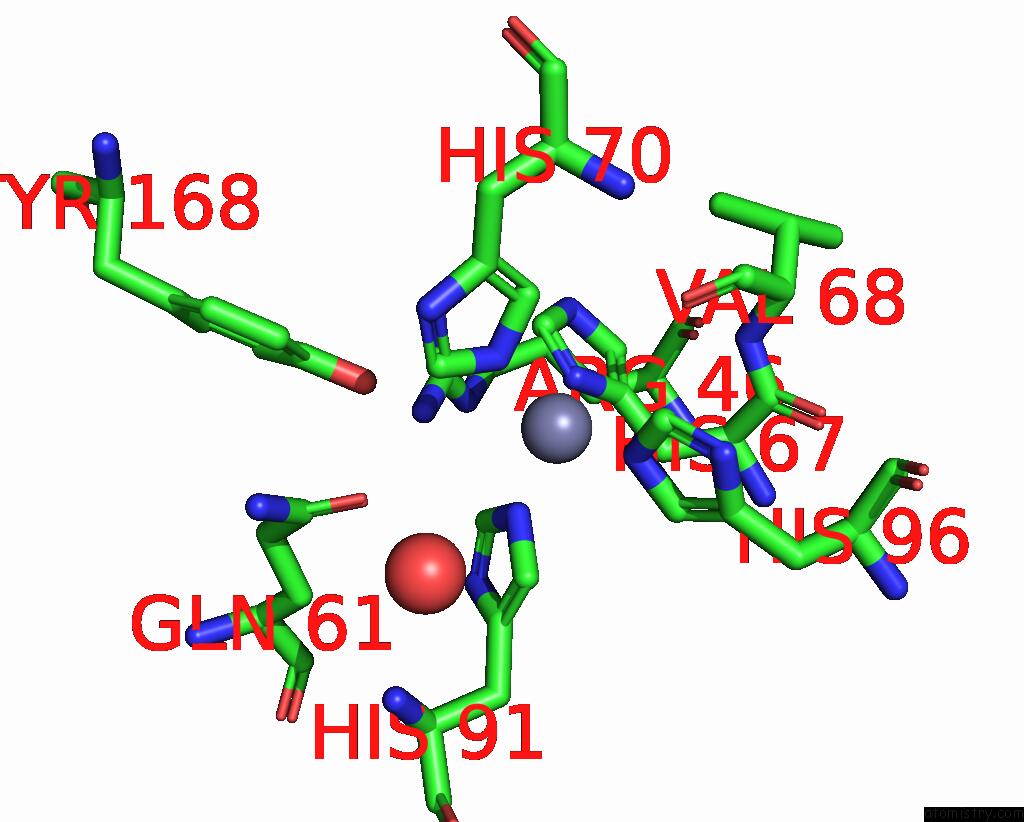
Mono view
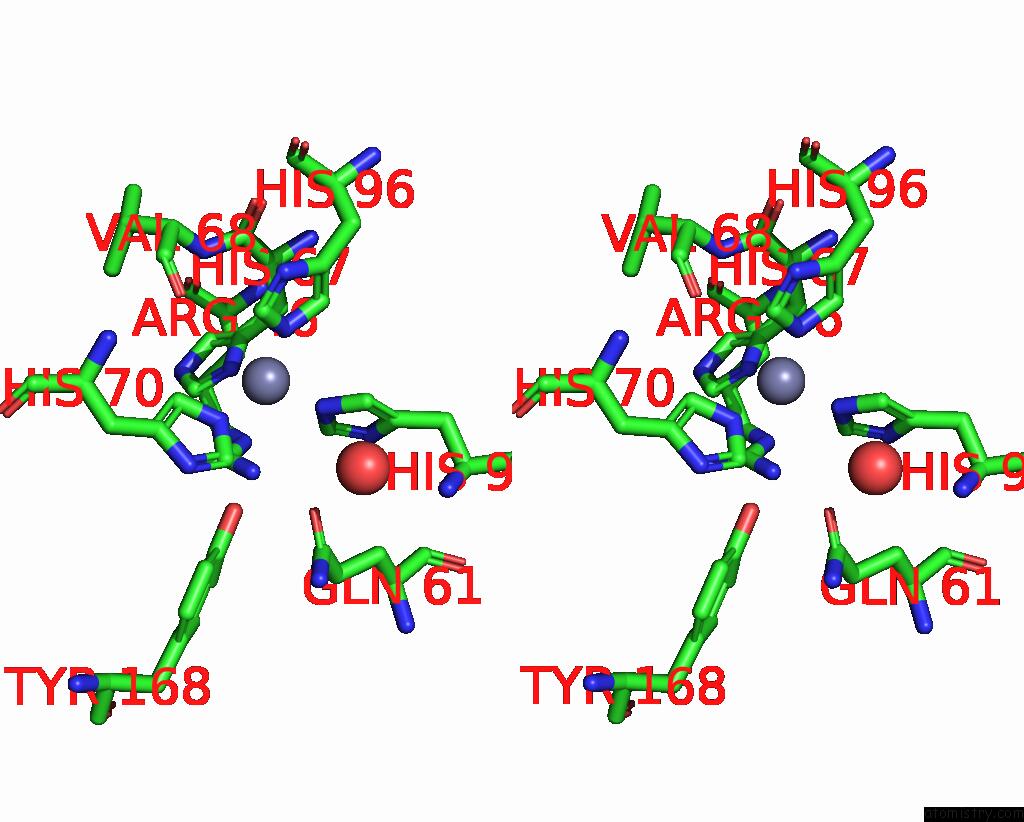
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 6 of Crystal Structures of Yrda From Escherichia Coli, A Homologous Protein of Gamma-Class Carbonic Anhydrase, Show Possible Allosteric Conformations within 5.0Å range:
|
Reference:
H.M.Park,
J.H.Park,
J.W.Choi,
J.E.Lee,
B.Y.Kim,
C.H.Jung,
J.S.Kim.
Structures of the Gamma-Class Carbonic Anhydrase Homologue Yrda Suggest A Possible Allosteric Switch Acta Crystallogr.,Sect.D V. 68 920 2012.
ISSN: ISSN 0907-4449
PubMed: 22868757
DOI: 10.1107/S0907444912017210
Page generated: Wed Aug 20 14:29:11 2025
ISSN: ISSN 0907-4449
PubMed: 22868757
DOI: 10.1107/S0907444912017210
Last articles
Zn in 4LIMZn in 4LI8
Zn in 4LI6
Zn in 4LIK
Zn in 4LIL
Zn in 4LGU
Zn in 4LH8
Zn in 4LGR
Zn in 4LHI
Zn in 4LFY