Zinc »
PDB 3lju-3lt9 »
3lka »
Zinc in PDB 3lka: Catalytic Domain of Human Mmp-12 Complexed with Hydroxamic Acid and Paramethoxy-Sulfonyl Amide
Enzymatic activity of Catalytic Domain of Human Mmp-12 Complexed with Hydroxamic Acid and Paramethoxy-Sulfonyl Amide
All present enzymatic activity of Catalytic Domain of Human Mmp-12 Complexed with Hydroxamic Acid and Paramethoxy-Sulfonyl Amide:
3.4.24.65;
3.4.24.65;
Protein crystallography data
The structure of Catalytic Domain of Human Mmp-12 Complexed with Hydroxamic Acid and Paramethoxy-Sulfonyl Amide, PDB code: 3lka
was solved by
V.Calderone,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 36.91 / 1.80 |
| Space group | C 1 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 51.566, 60.194, 54.215, 90.00, 115.09, 90.00 |
| R / Rfree (%) | 16.9 / 21 |
Other elements in 3lka:
The structure of Catalytic Domain of Human Mmp-12 Complexed with Hydroxamic Acid and Paramethoxy-Sulfonyl Amide also contains other interesting chemical elements:
| Calcium | (Ca) | 3 atoms |
Zinc Binding Sites:
The binding sites of Zinc atom in the Catalytic Domain of Human Mmp-12 Complexed with Hydroxamic Acid and Paramethoxy-Sulfonyl Amide
(pdb code 3lka). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total 2 binding sites of Zinc where determined in the Catalytic Domain of Human Mmp-12 Complexed with Hydroxamic Acid and Paramethoxy-Sulfonyl Amide, PDB code: 3lka:
Jump to Zinc binding site number: 1; 2;
In total 2 binding sites of Zinc where determined in the Catalytic Domain of Human Mmp-12 Complexed with Hydroxamic Acid and Paramethoxy-Sulfonyl Amide, PDB code: 3lka:
Jump to Zinc binding site number: 1; 2;
Zinc binding site 1 out of 2 in 3lka
Go back to
Zinc binding site 1 out
of 2 in the Catalytic Domain of Human Mmp-12 Complexed with Hydroxamic Acid and Paramethoxy-Sulfonyl Amide

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Catalytic Domain of Human Mmp-12 Complexed with Hydroxamic Acid and Paramethoxy-Sulfonyl Amide within 5.0Å range:
|
Zinc binding site 2 out of 2 in 3lka
Go back to
Zinc binding site 2 out
of 2 in the Catalytic Domain of Human Mmp-12 Complexed with Hydroxamic Acid and Paramethoxy-Sulfonyl Amide
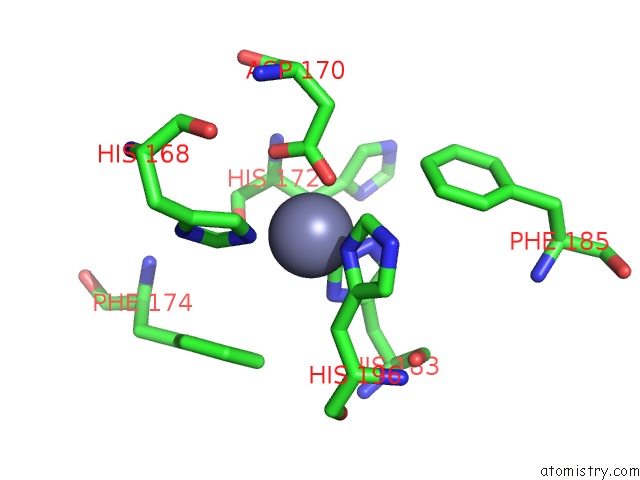
Mono view
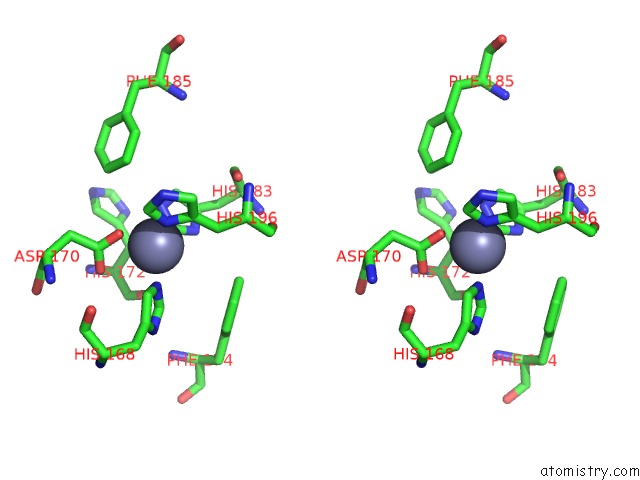
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 2 of Catalytic Domain of Human Mmp-12 Complexed with Hydroxamic Acid and Paramethoxy-Sulfonyl Amide within 5.0Å range:
|
Reference:
V.Borsi,
V.Calderone,
M.Fragai,
C.Luchinat,
N.Sarti.
Entropic Contribution to the Linking Coefficient in Fragment Based Drug Design: A Case Study. J.Med.Chem. V. 53 4285 2010.
ISSN: ISSN 0022-2623
PubMed: 20415416
DOI: 10.1021/JM901723Z
Page generated: Sat Oct 26 08:41:04 2024
ISSN: ISSN 0022-2623
PubMed: 20415416
DOI: 10.1021/JM901723Z
Last articles
K in 6ZKHK in 6ZKG
K in 6ZKF
K in 6ZKE
K in 6ZDP
K in 6ZKD
K in 6ZKC
K in 6ZGC
K in 6Z7V
K in 6ZK9