Zinc »
PDB 3iqx-3jr3 »
3jck »
Zinc in PDB 3jck: Structure of the Yeast 26S Proteasome Lid Sub-Complex
Enzymatic activity of Structure of the Yeast 26S Proteasome Lid Sub-Complex
All present enzymatic activity of Structure of the Yeast 26S Proteasome Lid Sub-Complex:
3.4.19.12;
3.4.19.12;
Zinc Binding Sites:
The binding sites of Zinc atom in the Structure of the Yeast 26S Proteasome Lid Sub-Complex
(pdb code 3jck). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total only one binding site of Zinc was determined in the Structure of the Yeast 26S Proteasome Lid Sub-Complex, PDB code: 3jck:
In total only one binding site of Zinc was determined in the Structure of the Yeast 26S Proteasome Lid Sub-Complex, PDB code: 3jck:
Zinc binding site 1 out of 1 in 3jck
Go back to
Zinc binding site 1 out
of 1 in the Structure of the Yeast 26S Proteasome Lid Sub-Complex
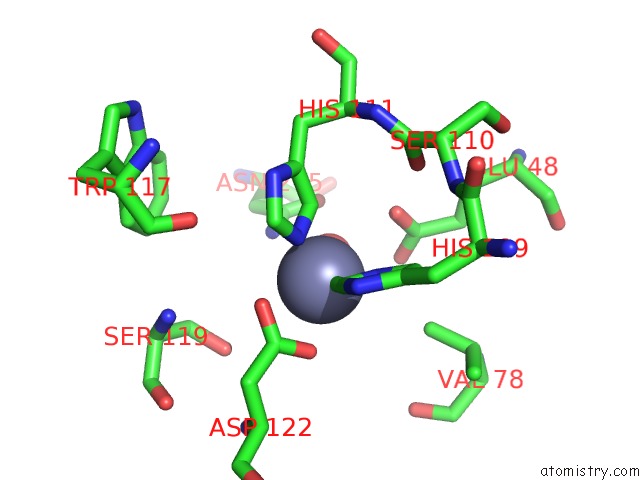
Mono view
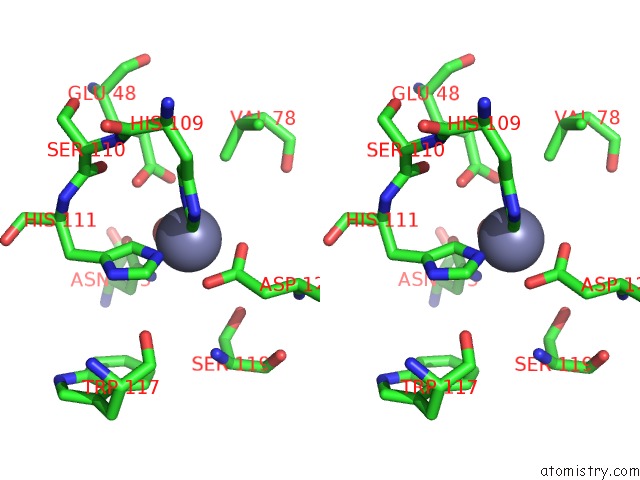
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Structure of the Yeast 26S Proteasome Lid Sub-Complex within 5.0Å range:
|
Reference:
C.M.Dambacher,
E.J.Worden,
M.A.Herzik,
A.Martin,
G.C.Lander.
Atomic Structure of the 26S Proteasome Lid Reveals the Mechanism of Deubiquitinase Inhibition. Elife V. 5 13027 2016.
ISSN: ESSN 2050-084X
PubMed: 26744777
DOI: 10.7554/ELIFE.13027
Page generated: Sat Oct 26 07:28:53 2024
ISSN: ESSN 2050-084X
PubMed: 26744777
DOI: 10.7554/ELIFE.13027
Last articles
Na in 9IN7Na in 9IKG
Na in 9IKH
Na in 9IKF
Na in 9IGK
Na in 9IGL
Na in 9ICY
Na in 9ICX
Na in 9ICW
Na in 9ICV