Zinc »
PDB 3f7u-3fl2 »
3fav »
Zinc in PDB 3fav: Structure of the CFP10-ESAT6 Complex From Mycobacterium Tuberculosis
Protein crystallography data
The structure of Structure of the CFP10-ESAT6 Complex From Mycobacterium Tuberculosis, PDB code: 3fav
was solved by
C.Poulsen,
S.J.Holton,
M.Wilmanns,
Y.H.Song,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 19.86 / 2.15 |
| Space group | C 1 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 160.340, 23.930, 83.860, 90.00, 94.36, 90.00 |
| R / Rfree (%) | 19.9 / 23.3 |
Zinc Binding Sites:
The binding sites of Zinc atom in the Structure of the CFP10-ESAT6 Complex From Mycobacterium Tuberculosis
(pdb code 3fav). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total 7 binding sites of Zinc where determined in the Structure of the CFP10-ESAT6 Complex From Mycobacterium Tuberculosis, PDB code: 3fav:
Jump to Zinc binding site number: 1; 2; 3; 4; 5; 6; 7;
In total 7 binding sites of Zinc where determined in the Structure of the CFP10-ESAT6 Complex From Mycobacterium Tuberculosis, PDB code: 3fav:
Jump to Zinc binding site number: 1; 2; 3; 4; 5; 6; 7;
Zinc binding site 1 out of 7 in 3fav
Go back to
Zinc binding site 1 out
of 7 in the Structure of the CFP10-ESAT6 Complex From Mycobacterium Tuberculosis
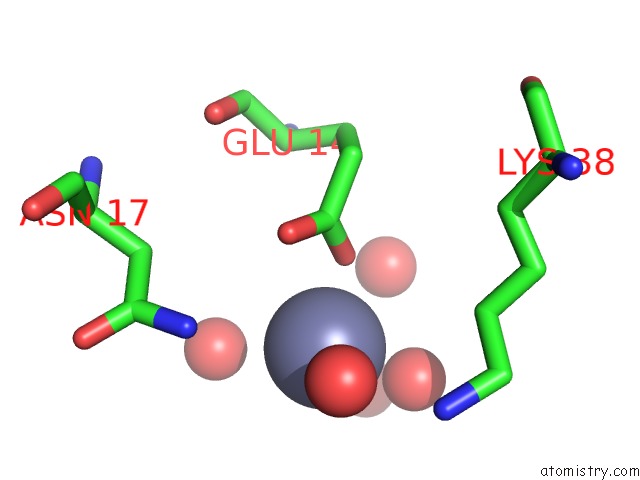
Mono view
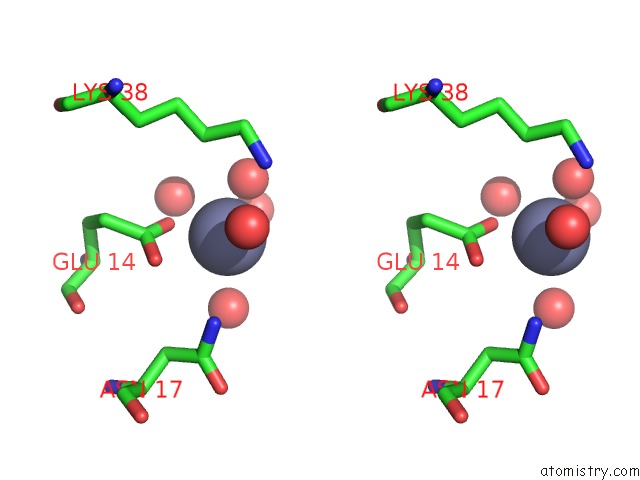
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Structure of the CFP10-ESAT6 Complex From Mycobacterium Tuberculosis within 5.0Å range:
|
Zinc binding site 2 out of 7 in 3fav
Go back to
Zinc binding site 2 out
of 7 in the Structure of the CFP10-ESAT6 Complex From Mycobacterium Tuberculosis
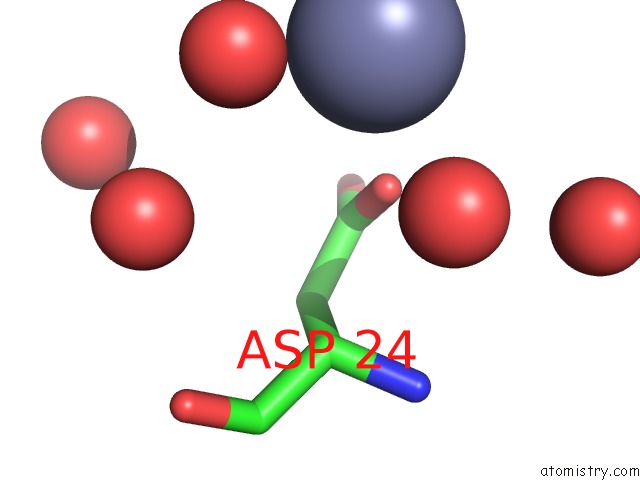
Mono view
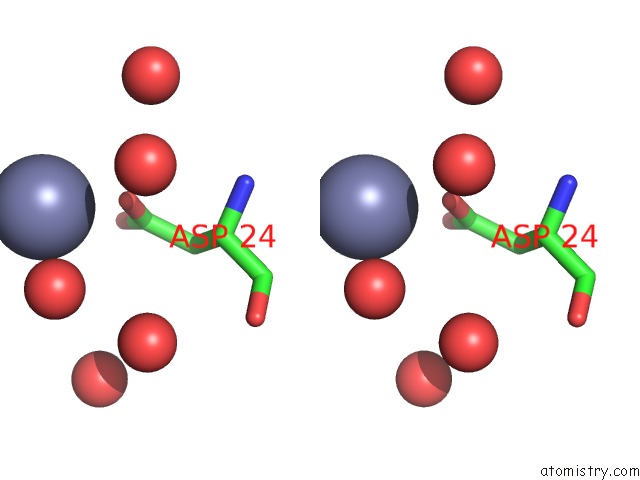
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 2 of Structure of the CFP10-ESAT6 Complex From Mycobacterium Tuberculosis within 5.0Å range:
|
Zinc binding site 3 out of 7 in 3fav
Go back to
Zinc binding site 3 out
of 7 in the Structure of the CFP10-ESAT6 Complex From Mycobacterium Tuberculosis
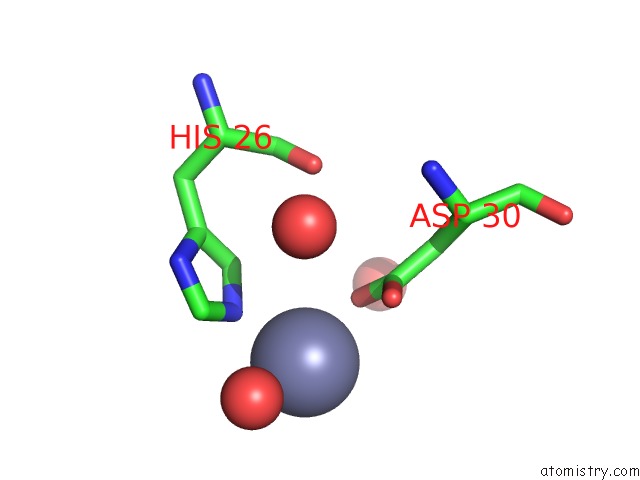
Mono view
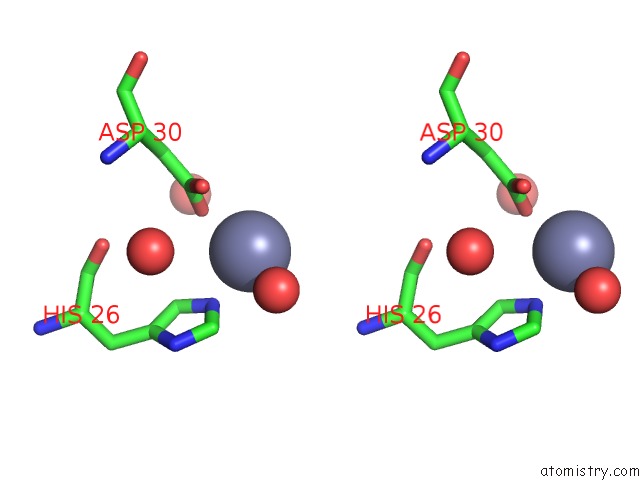
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 3 of Structure of the CFP10-ESAT6 Complex From Mycobacterium Tuberculosis within 5.0Å range:
|
Zinc binding site 4 out of 7 in 3fav
Go back to
Zinc binding site 4 out
of 7 in the Structure of the CFP10-ESAT6 Complex From Mycobacterium Tuberculosis
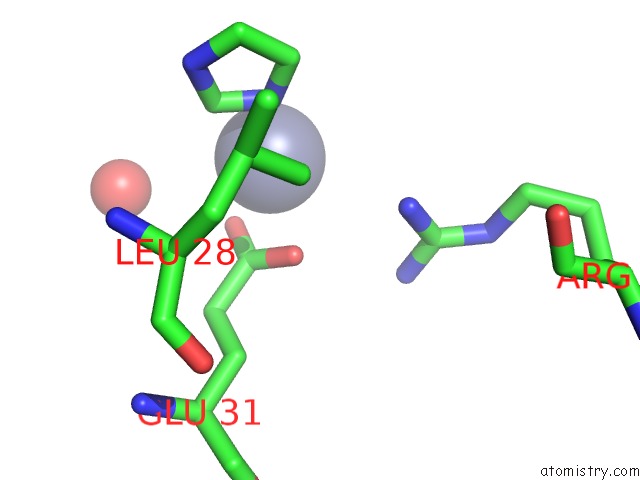
Mono view
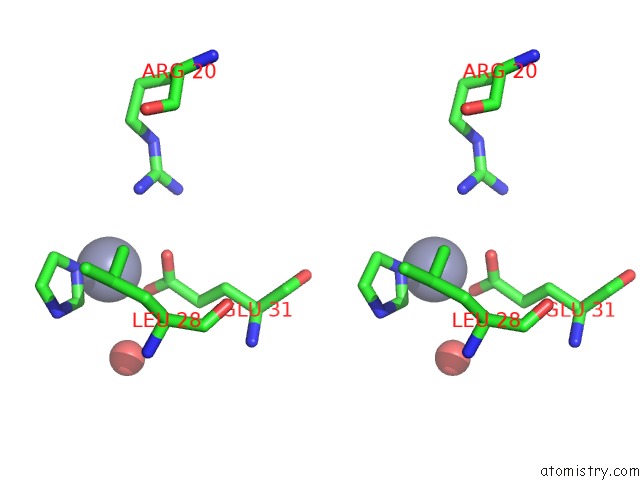
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 4 of Structure of the CFP10-ESAT6 Complex From Mycobacterium Tuberculosis within 5.0Å range:
|
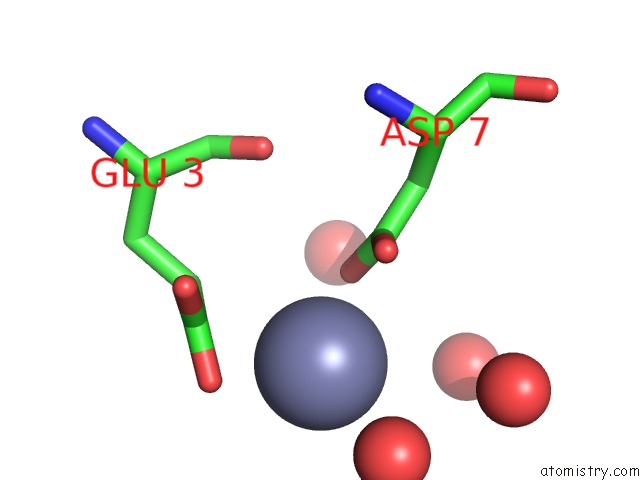
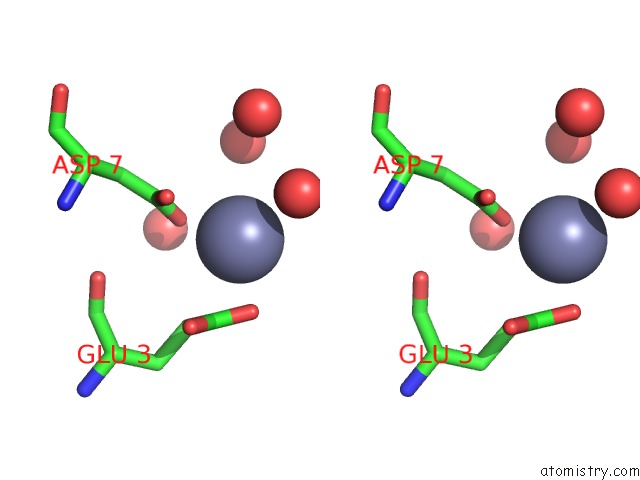
Zinc binding site 5 out of 7 in 3fav
Go back to
Zinc binding site 5 out
of 7 in the Structure of the CFP10-ESAT6 Complex From Mycobacterium Tuberculosis
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 5 of Structure of the CFP10-ESAT6 Complex From Mycobacterium Tuberculosis within 5.0Å range:
|
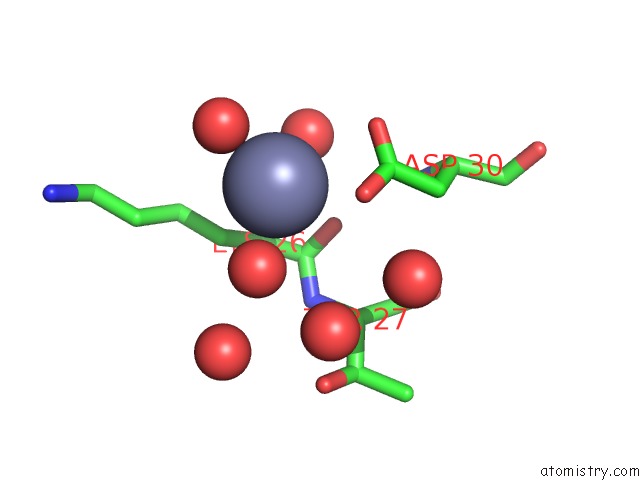
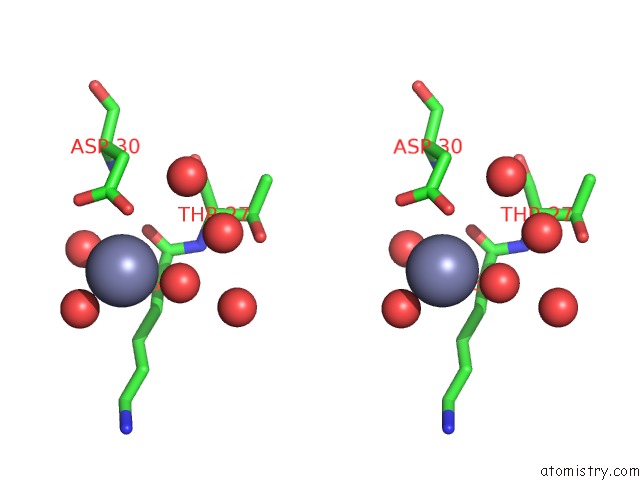
Zinc binding site 6 out of 7 in 3fav
Go back to
Zinc binding site 6 out
of 7 in the Structure of the CFP10-ESAT6 Complex From Mycobacterium Tuberculosis
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 6 of Structure of the CFP10-ESAT6 Complex From Mycobacterium Tuberculosis within 5.0Å range:
|
Zinc binding site 7 out of 7 in 3fav
Go back to
Zinc binding site 7 out
of 7 in the Structure of the CFP10-ESAT6 Complex From Mycobacterium Tuberculosis
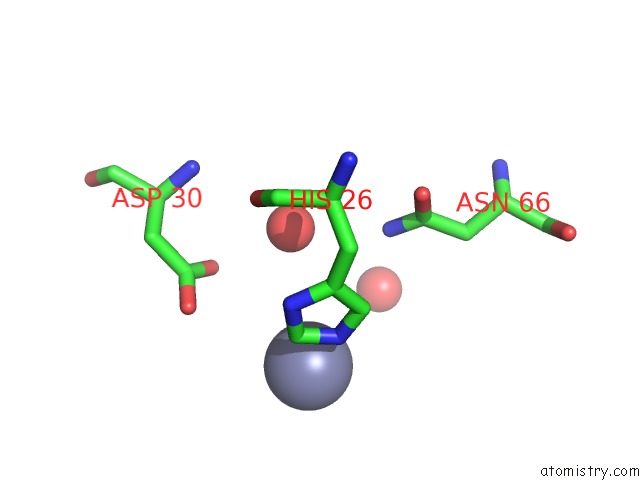
Mono view
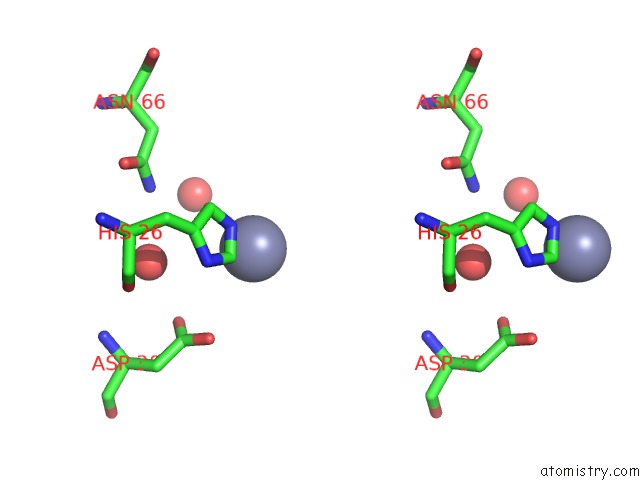
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 7 of Structure of the CFP10-ESAT6 Complex From Mycobacterium Tuberculosis within 5.0Å range:
|
Reference:
C.Poulsen,
S.Panjikar,
S.J.Holton,
M.Wilmanns,
Y.H.Song.
WXG100 Protein Superfamily Consists of Three Subfamilies and Exhibits An Alpha-Helical C-Terminal Conserved Residue Pattern. Plos One V. 9 89313 2014.
ISSN: ESSN 1932-6203
PubMed: 24586681
DOI: 10.1371/JOURNAL.PONE.0089313
Page generated: Thu Oct 24 13:06:25 2024
ISSN: ESSN 1932-6203
PubMed: 24586681
DOI: 10.1371/JOURNAL.PONE.0089313
Last articles
Mg in 3LD0Mg in 3LLU
Mg in 3LK9
Mg in 3LJ0
Mg in 3LDW
Mg in 3LIJ
Mg in 3LIE
Mg in 3LGZ
Mg in 3LHR
Mg in 3LGY