Zinc »
PDB 3cjp-3czs »
3cqj »
Zinc in PDB 3cqj: Crystal Structure of L-Xylulose-5-Phosphate 3-Epimerase Ulae (Form B) Complex with ZN2+
Enzymatic activity of Crystal Structure of L-Xylulose-5-Phosphate 3-Epimerase Ulae (Form B) Complex with ZN2+
All present enzymatic activity of Crystal Structure of L-Xylulose-5-Phosphate 3-Epimerase Ulae (Form B) Complex with ZN2+:
5.1.3.22;
5.1.3.22;
Protein crystallography data
The structure of Crystal Structure of L-Xylulose-5-Phosphate 3-Epimerase Ulae (Form B) Complex with ZN2+, PDB code: 3cqj
was solved by
R.Shi,
A.Matte,
M.Cygler,
Montreal-Kingston Bacterial Structuralgenomics Initiative (Bsgi),
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 50.00 / 2.04 |
| Space group | C 2 2 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 104.208, 132.596, 81.798, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 18.3 / 21.3 |
Zinc Binding Sites:
The binding sites of Zinc atom in the Crystal Structure of L-Xylulose-5-Phosphate 3-Epimerase Ulae (Form B) Complex with ZN2+
(pdb code 3cqj). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total 4 binding sites of Zinc where determined in the Crystal Structure of L-Xylulose-5-Phosphate 3-Epimerase Ulae (Form B) Complex with ZN2+, PDB code: 3cqj:
Jump to Zinc binding site number: 1; 2; 3; 4;
In total 4 binding sites of Zinc where determined in the Crystal Structure of L-Xylulose-5-Phosphate 3-Epimerase Ulae (Form B) Complex with ZN2+, PDB code: 3cqj:
Jump to Zinc binding site number: 1; 2; 3; 4;
Zinc binding site 1 out of 4 in 3cqj
Go back to
Zinc binding site 1 out
of 4 in the Crystal Structure of L-Xylulose-5-Phosphate 3-Epimerase Ulae (Form B) Complex with ZN2+
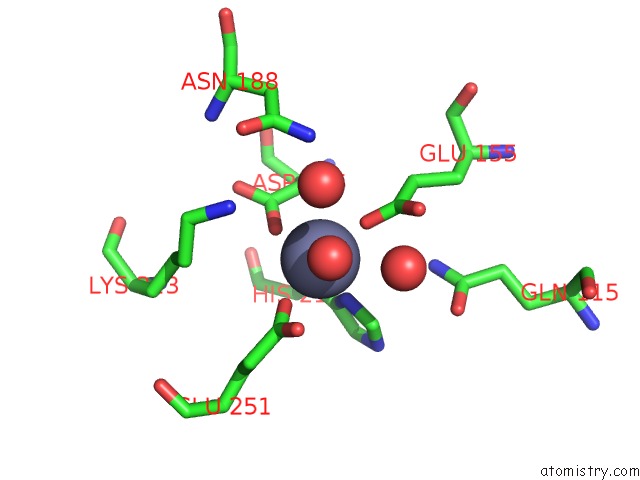
Mono view
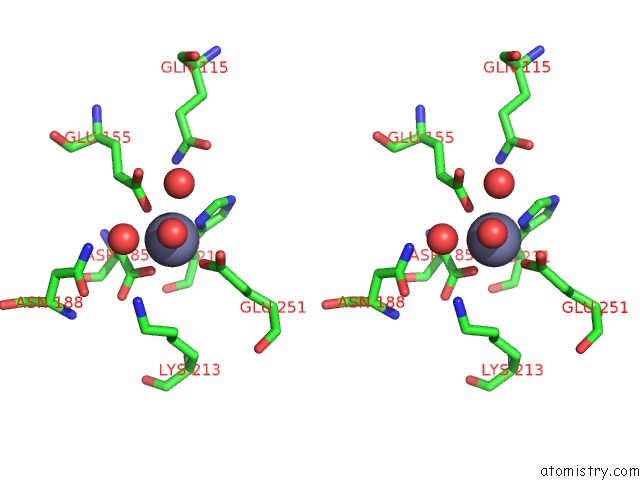
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Crystal Structure of L-Xylulose-5-Phosphate 3-Epimerase Ulae (Form B) Complex with ZN2+ within 5.0Å range:
|
Zinc binding site 2 out of 4 in 3cqj
Go back to
Zinc binding site 2 out
of 4 in the Crystal Structure of L-Xylulose-5-Phosphate 3-Epimerase Ulae (Form B) Complex with ZN2+
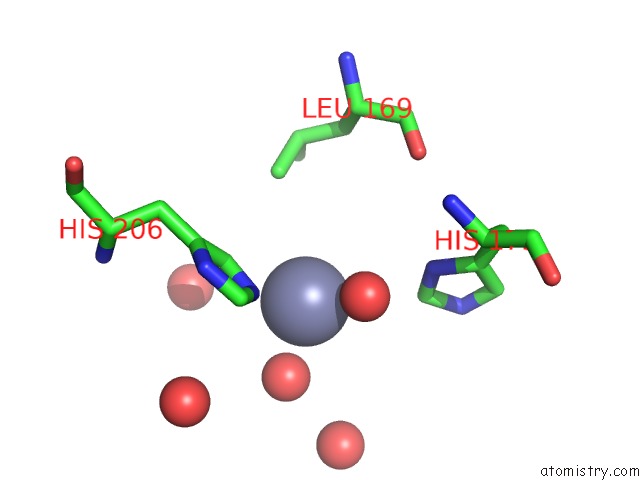
Mono view
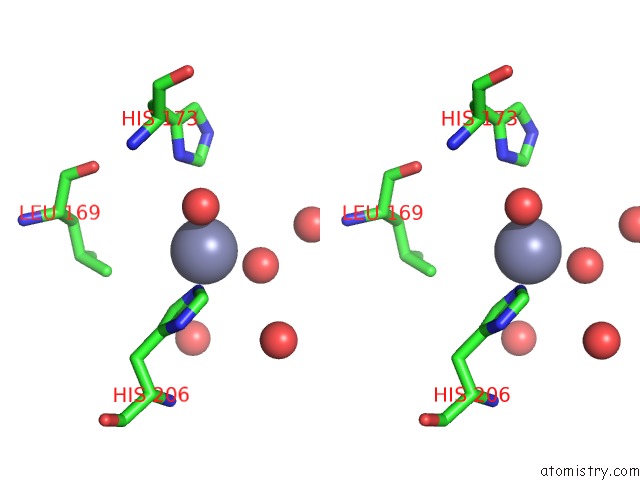
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 2 of Crystal Structure of L-Xylulose-5-Phosphate 3-Epimerase Ulae (Form B) Complex with ZN2+ within 5.0Å range:
|
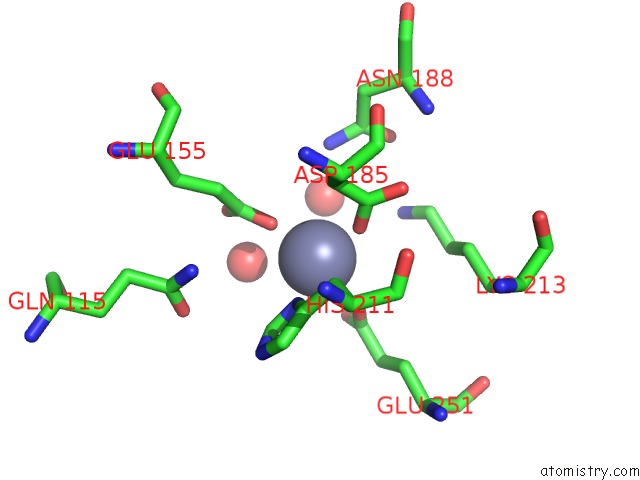
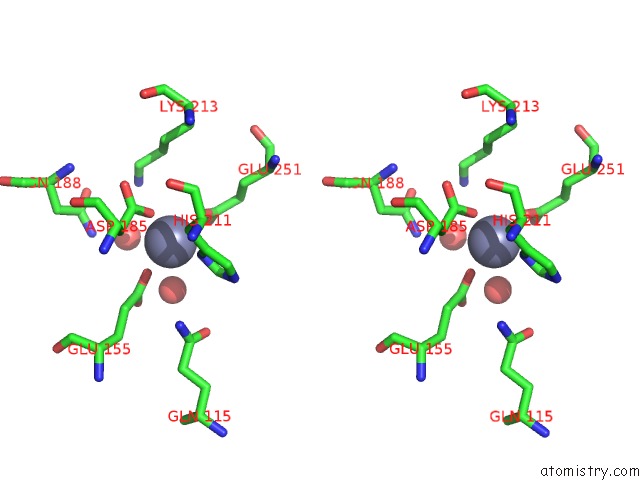
Zinc binding site 3 out of 4 in 3cqj
Go back to
Zinc binding site 3 out
of 4 in the Crystal Structure of L-Xylulose-5-Phosphate 3-Epimerase Ulae (Form B) Complex with ZN2+
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 3 of Crystal Structure of L-Xylulose-5-Phosphate 3-Epimerase Ulae (Form B) Complex with ZN2+ within 5.0Å range:
|
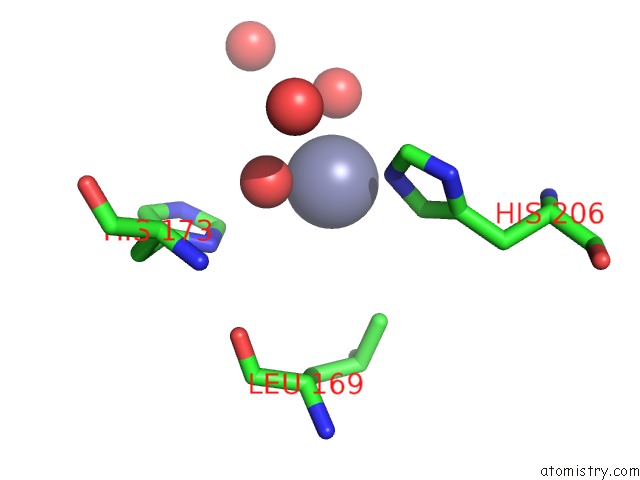
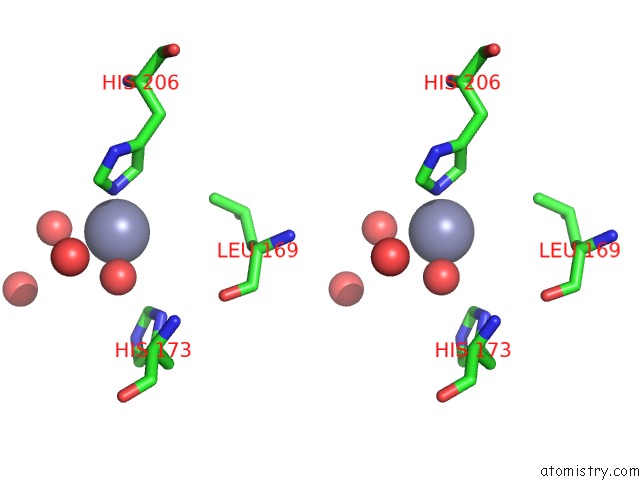
Zinc binding site 4 out of 4 in 3cqj
Go back to
Zinc binding site 4 out
of 4 in the Crystal Structure of L-Xylulose-5-Phosphate 3-Epimerase Ulae (Form B) Complex with ZN2+
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 4 of Crystal Structure of L-Xylulose-5-Phosphate 3-Epimerase Ulae (Form B) Complex with ZN2+ within 5.0Å range:
|
Reference:
R.Shi,
M.Pineda,
E.Ajamian,
Q.Cui,
A.Matte,
M.Cygler.
Structure of L-Xylulose-5-Phosphate 3-Epimerase (Ulae) From the Anaerobic L-Ascorbate Utilization Pathway of Escherichia Coli: Identification of A Novel Phosphate Binding Motif Within A Tim Barrel Fold. J.Bacteriol. V. 190 8137 2008.
ISSN: ISSN 0021-9193
PubMed: 18849419
DOI: 10.1128/JB.01049-08
Page generated: Thu Oct 24 11:54:45 2024
ISSN: ISSN 0021-9193
PubMed: 18849419
DOI: 10.1128/JB.01049-08
Last articles
K in 7G36K in 7G34
K in 7G30
K in 7G2Y
K in 7G2X
K in 7G2W
K in 7G2V
K in 7G2U
K in 7G2T
K in 7G2S