Zinc »
PDB 3adr-3avs »
3al0 »
Zinc in PDB 3al0: Crystal Structure of the Glutamine Transamidosome From Thermotoga Maritima in the Glutamylation State.
Enzymatic activity of Crystal Structure of the Glutamine Transamidosome From Thermotoga Maritima in the Glutamylation State.
All present enzymatic activity of Crystal Structure of the Glutamine Transamidosome From Thermotoga Maritima in the Glutamylation State.:
6.1.1.17; 6.3.5.7;
6.1.1.17; 6.3.5.7;
Protein crystallography data
The structure of Crystal Structure of the Glutamine Transamidosome From Thermotoga Maritima in the Glutamylation State., PDB code: 3al0
was solved by
T.Ito,
S.Yokoyama,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 48.67 / 3.37 |
| Space group | P 2 2 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 76.954, 125.661, 313.386, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 19.5 / 26.9 |
Zinc Binding Sites:
The binding sites of Zinc atom in the Crystal Structure of the Glutamine Transamidosome From Thermotoga Maritima in the Glutamylation State.
(pdb code 3al0). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total only one binding site of Zinc was determined in the Crystal Structure of the Glutamine Transamidosome From Thermotoga Maritima in the Glutamylation State., PDB code: 3al0:
In total only one binding site of Zinc was determined in the Crystal Structure of the Glutamine Transamidosome From Thermotoga Maritima in the Glutamylation State., PDB code: 3al0:
Zinc binding site 1 out of 1 in 3al0
Go back to
Zinc binding site 1 out
of 1 in the Crystal Structure of the Glutamine Transamidosome From Thermotoga Maritima in the Glutamylation State.
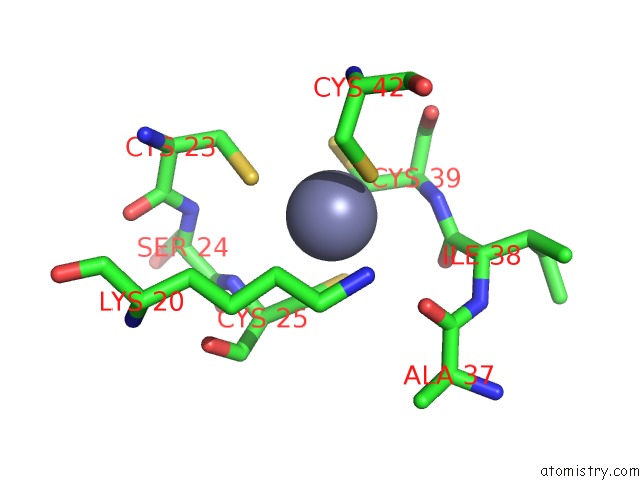
Mono view
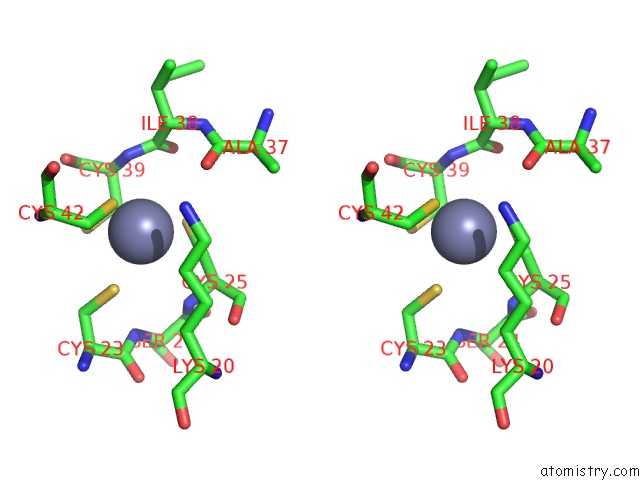
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Crystal Structure of the Glutamine Transamidosome From Thermotoga Maritima in the Glutamylation State. within 5.0Å range:
|
Reference:
T.Ito,
S.Yokoyama.
Two Enzymes Bound to One Transfer Rna Assume Alternative Conformations For Consecutive Reactions. Nature V. 467 612 2010.
ISSN: ISSN 0028-0836
PubMed: 20882017
DOI: 10.1038/NATURE09411
Page generated: Thu Oct 24 11:13:01 2024
ISSN: ISSN 0028-0836
PubMed: 20882017
DOI: 10.1038/NATURE09411
Last articles
Mg in 6NMCMg in 6NL5
Mg in 6NMA
Mg in 6NLB
Mg in 6NL9
Mg in 6NL0
Mg in 6NKZ
Mg in 6NKW
Mg in 6NKY
Mg in 6NKX