Zinc »
PDB 2mmk-2naa »
2mqv »
Zinc in PDB 2mqv: Solution uc(Nmr) Structure of the U5-Primer Binding Site (U5-Pbs) Domain of Murine Leukemia Virus Rna Genome Bound to the Retroviral Nucleocapsid Protein
Zinc Binding Sites:
The binding sites of Zinc atom in the Solution uc(Nmr) Structure of the U5-Primer Binding Site (U5-Pbs) Domain of Murine Leukemia Virus Rna Genome Bound to the Retroviral Nucleocapsid Protein
(pdb code 2mqv). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total only one binding site of Zinc was determined in the Solution uc(Nmr) Structure of the U5-Primer Binding Site (U5-Pbs) Domain of Murine Leukemia Virus Rna Genome Bound to the Retroviral Nucleocapsid Protein, PDB code: 2mqv:
In total only one binding site of Zinc was determined in the Solution uc(Nmr) Structure of the U5-Primer Binding Site (U5-Pbs) Domain of Murine Leukemia Virus Rna Genome Bound to the Retroviral Nucleocapsid Protein, PDB code: 2mqv:
Zinc binding site 1 out of 1 in 2mqv
Go back to
Zinc binding site 1 out
of 1 in the Solution uc(Nmr) Structure of the U5-Primer Binding Site (U5-Pbs) Domain of Murine Leukemia Virus Rna Genome Bound to the Retroviral Nucleocapsid Protein
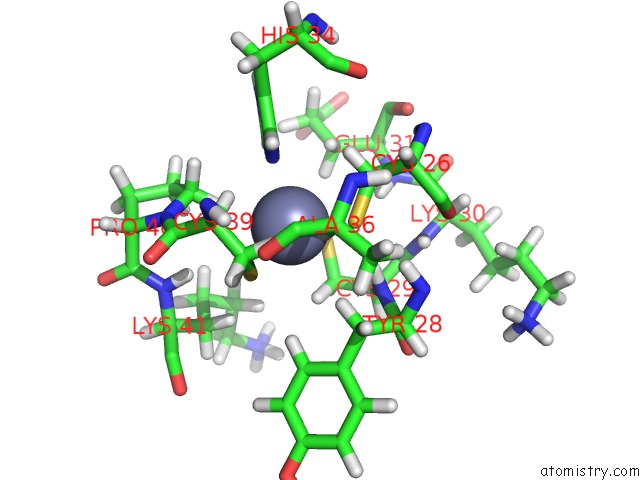
Mono view
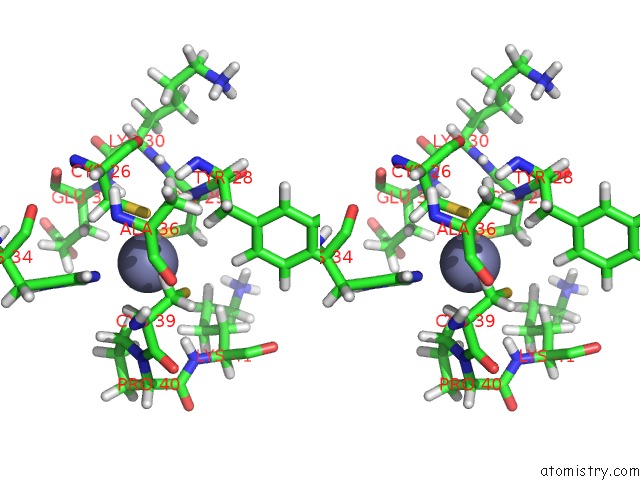
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Solution uc(Nmr) Structure of the U5-Primer Binding Site (U5-Pbs) Domain of Murine Leukemia Virus Rna Genome Bound to the Retroviral Nucleocapsid Protein within 5.0Å range:
|
Reference:
S.B.Miller,
F.Z.Yildiz,
J.A.Lo,
B.Wang,
V.M.D'souza.
A Structure-Based Mechanism For Trna and Retroviral Rna Remodelling During Primer Annealing. Nature 2014.
ISSN: ESSN 1476-4687
PubMed: 25209668
DOI: 10.1038/NATURE13709
Page generated: Wed Aug 20 04:26:52 2025
ISSN: ESSN 1476-4687
PubMed: 25209668
DOI: 10.1038/NATURE13709
Last articles
Zn in 3F0DZn in 3F0F
Zn in 3F07
Zn in 3F0E
Zn in 3EZ5
Zn in 3F06
Zn in 3EZT
Zn in 3EZP
Zn in 3EYX
Zn in 3EYY