Zinc »
PDB 2kzz-2lgl »
2lgg »
Zinc in PDB 2lgg: Structure of Phd Domain of UHRF1 in Complex with H3 Peptide
Zinc Binding Sites:
The binding sites of Zinc atom in the Structure of Phd Domain of UHRF1 in Complex with H3 Peptide
(pdb code 2lgg). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total 3 binding sites of Zinc where determined in the Structure of Phd Domain of UHRF1 in Complex with H3 Peptide, PDB code: 2lgg:
Jump to Zinc binding site number: 1; 2; 3;
In total 3 binding sites of Zinc where determined in the Structure of Phd Domain of UHRF1 in Complex with H3 Peptide, PDB code: 2lgg:
Jump to Zinc binding site number: 1; 2; 3;
Zinc binding site 1 out of 3 in 2lgg
Go back to
Zinc binding site 1 out
of 3 in the Structure of Phd Domain of UHRF1 in Complex with H3 Peptide
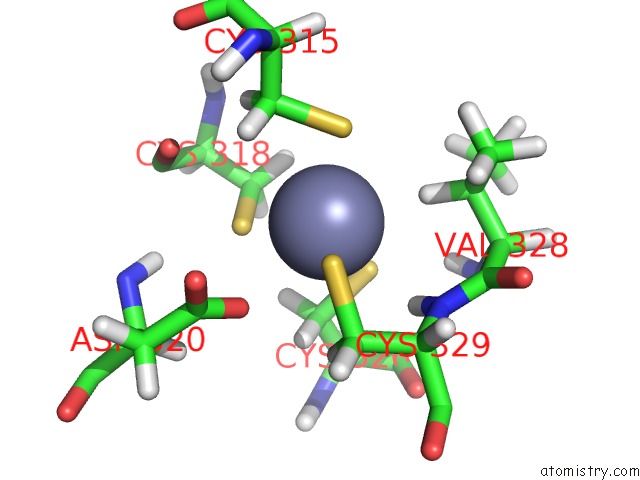
Mono view
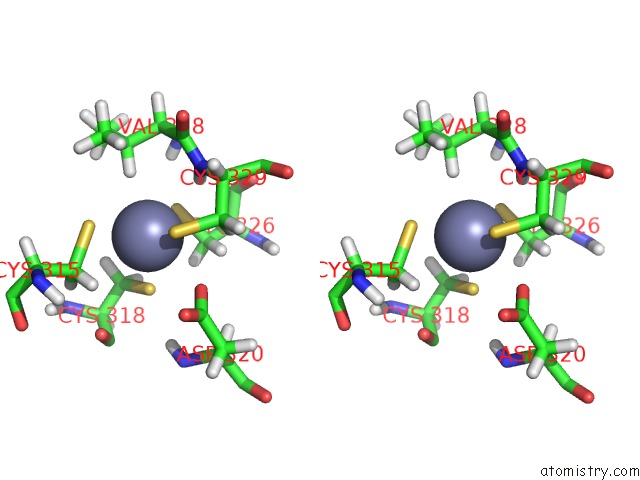
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Structure of Phd Domain of UHRF1 in Complex with H3 Peptide within 5.0Å range:
|
Zinc binding site 2 out of 3 in 2lgg
Go back to
Zinc binding site 2 out
of 3 in the Structure of Phd Domain of UHRF1 in Complex with H3 Peptide
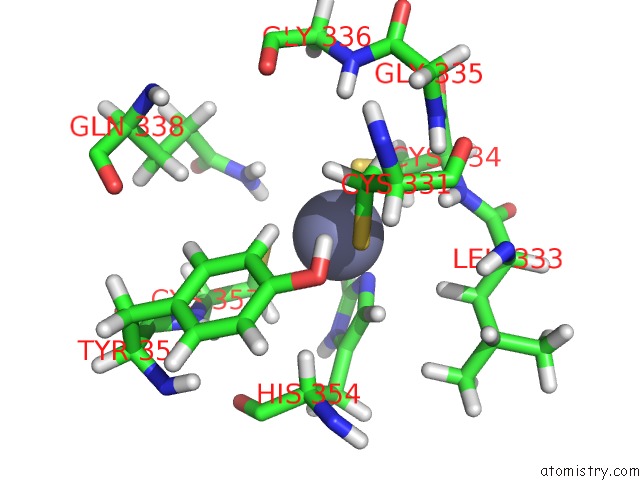
Mono view
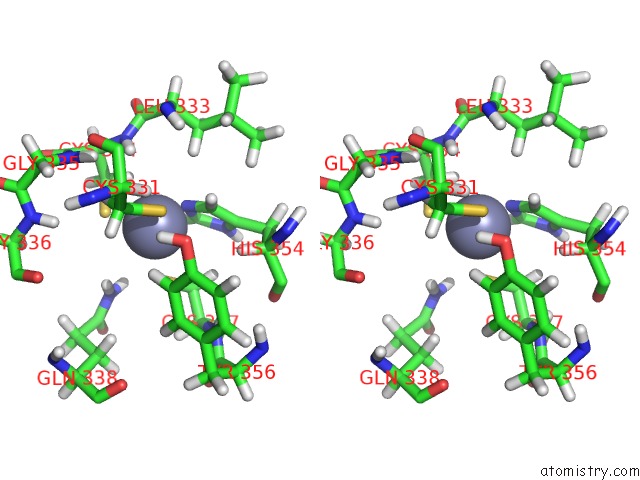
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 2 of Structure of Phd Domain of UHRF1 in Complex with H3 Peptide within 5.0Å range:
|
Zinc binding site 3 out of 3 in 2lgg
Go back to
Zinc binding site 3 out
of 3 in the Structure of Phd Domain of UHRF1 in Complex with H3 Peptide
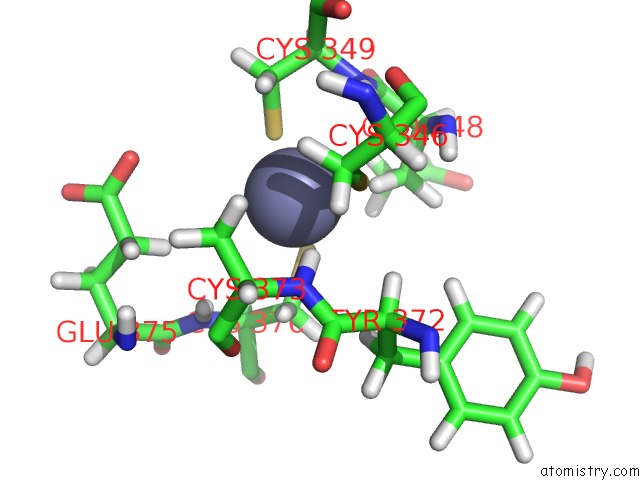
Mono view
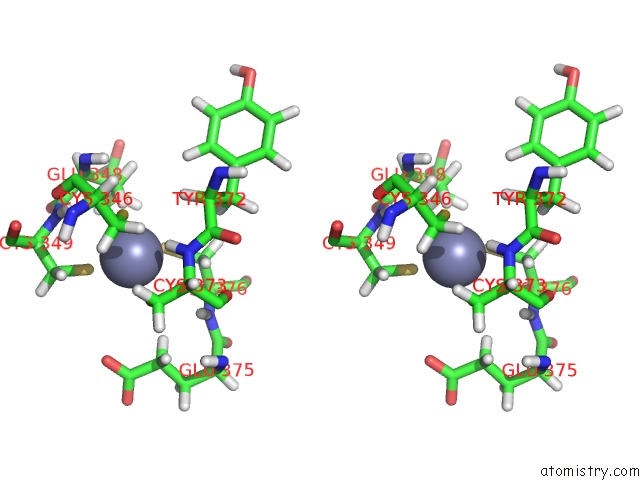
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 3 of Structure of Phd Domain of UHRF1 in Complex with H3 Peptide within 5.0Å range:
|
Reference:
C.Wang,
J.Shen,
Z.Yang,
P.Chen,
B.Zhao,
W.Hu,
W.Lan,
X.Tong,
H.Wu,
G.Li,
C.Cao.
Structural Basis For Site-Specific Reading of Unmodified R2 of Histone H3 Tail By UHRF1 Phd Finger. Cell Res. V. 21 1379 2011.
ISSN: ISSN 1001-0602
PubMed: 21808299
DOI: 10.1038/CR.2011.123
Page generated: Thu Oct 17 01:49:45 2024
ISSN: ISSN 1001-0602
PubMed: 21808299
DOI: 10.1038/CR.2011.123
Last articles
Fe in 2YXOFe in 2YRS
Fe in 2YXC
Fe in 2YNM
Fe in 2YVJ
Fe in 2YP1
Fe in 2YU2
Fe in 2YU1
Fe in 2YQB
Fe in 2YOO