Zinc »
PDB 2ics-2iv0 »
2ida »
Zinc in PDB 2ida: Solution uc(Nmr) Structure of Protein RPA1320 From Rhodopseudomonas Palustris. Northeast Structural Genomics Consortium Target RPT3; Ontario Center For Structural Proteomics Target RP1313.
Zinc Binding Sites:
The binding sites of Zinc atom in the Solution uc(Nmr) Structure of Protein RPA1320 From Rhodopseudomonas Palustris. Northeast Structural Genomics Consortium Target RPT3; Ontario Center For Structural Proteomics Target RP1313.
(pdb code 2ida). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total 2 binding sites of Zinc where determined in the Solution uc(Nmr) Structure of Protein RPA1320 From Rhodopseudomonas Palustris. Northeast Structural Genomics Consortium Target RPT3; Ontario Center For Structural Proteomics Target RP1313., PDB code: 2ida:
Jump to Zinc binding site number: 1; 2;
In total 2 binding sites of Zinc where determined in the Solution uc(Nmr) Structure of Protein RPA1320 From Rhodopseudomonas Palustris. Northeast Structural Genomics Consortium Target RPT3; Ontario Center For Structural Proteomics Target RP1313., PDB code: 2ida:
Jump to Zinc binding site number: 1; 2;
Zinc binding site 1 out of 2 in 2ida
Go back to
Zinc binding site 1 out
of 2 in the Solution uc(Nmr) Structure of Protein RPA1320 From Rhodopseudomonas Palustris. Northeast Structural Genomics Consortium Target RPT3; Ontario Center For Structural Proteomics Target RP1313.
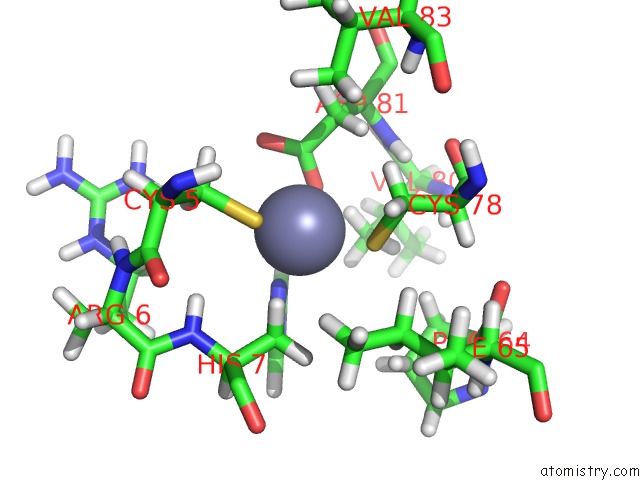
Mono view
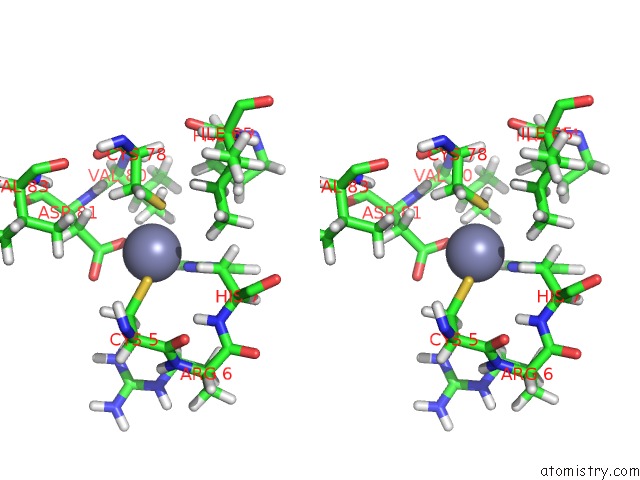
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Solution uc(Nmr) Structure of Protein RPA1320 From Rhodopseudomonas Palustris. Northeast Structural Genomics Consortium Target RPT3; Ontario Center For Structural Proteomics Target RP1313. within 5.0Å range:
|
Zinc binding site 2 out of 2 in 2ida
Go back to
Zinc binding site 2 out
of 2 in the Solution uc(Nmr) Structure of Protein RPA1320 From Rhodopseudomonas Palustris. Northeast Structural Genomics Consortium Target RPT3; Ontario Center For Structural Proteomics Target RP1313.
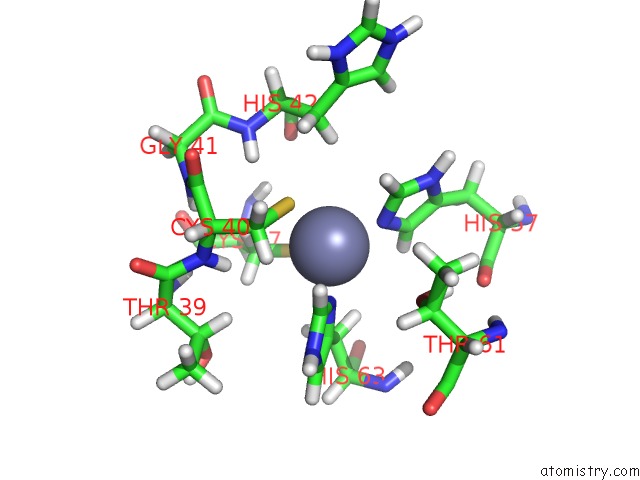
Mono view
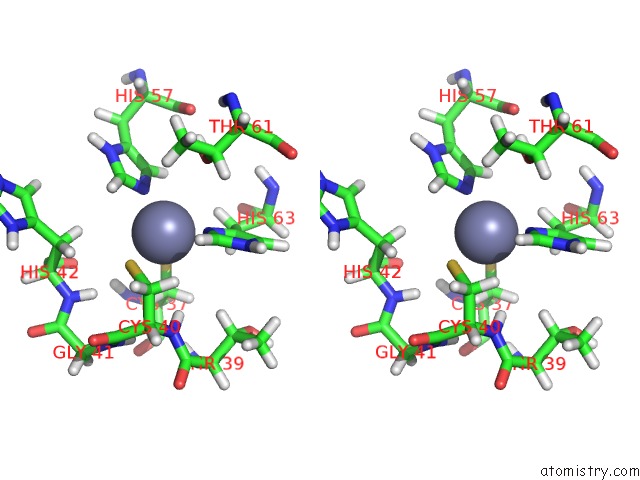
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 2 of Solution uc(Nmr) Structure of Protein RPA1320 From Rhodopseudomonas Palustris. Northeast Structural Genomics Consortium Target RPT3; Ontario Center For Structural Proteomics Target RP1313. within 5.0Å range:
|
Reference:
A.Lemak,
A.Yee,
J.A.Lukin,
M.Karra,
A.Gutmanas,
C.H.Arrowsmith.
Solution Structure of RPA1320 To Be Published.
Page generated: Thu Oct 17 00:53:25 2024
Last articles
Mg in 6LU1Mg in 6LT4
Mg in 6LY7
Mg in 6LY6
Mg in 6LY3
Mg in 6LX1
Mg in 6LTW
Mg in 6LVW
Mg in 6LUH
Mg in 6LTS