Zinc »
PDB 2ctt-2das »
2cuq »
Zinc in PDB 2cuq: Solution Structure of Second Lim Domain From Human Skeletal Muscle Lim-Protein 2
Zinc Binding Sites:
The binding sites of Zinc atom in the Solution Structure of Second Lim Domain From Human Skeletal Muscle Lim-Protein 2
(pdb code 2cuq). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total 2 binding sites of Zinc where determined in the Solution Structure of Second Lim Domain From Human Skeletal Muscle Lim-Protein 2, PDB code: 2cuq:
Jump to Zinc binding site number: 1; 2;
In total 2 binding sites of Zinc where determined in the Solution Structure of Second Lim Domain From Human Skeletal Muscle Lim-Protein 2, PDB code: 2cuq:
Jump to Zinc binding site number: 1; 2;
Zinc binding site 1 out of 2 in 2cuq
Go back to
Zinc binding site 1 out
of 2 in the Solution Structure of Second Lim Domain From Human Skeletal Muscle Lim-Protein 2
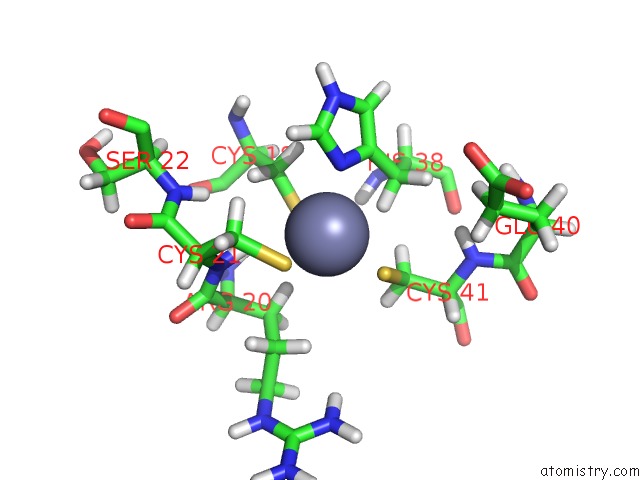
Mono view
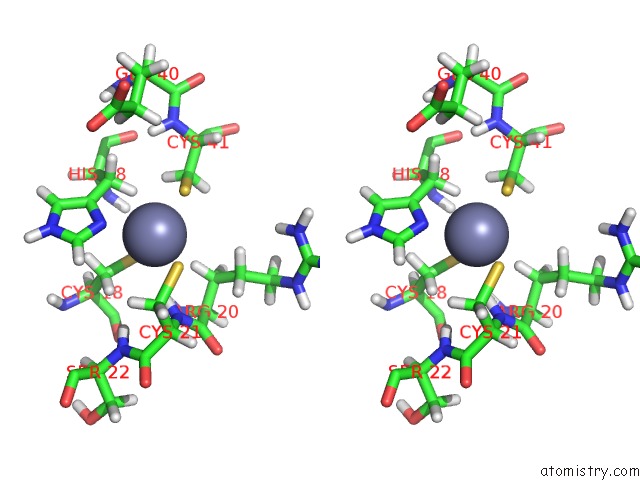
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Solution Structure of Second Lim Domain From Human Skeletal Muscle Lim-Protein 2 within 5.0Å range:
|
Zinc binding site 2 out of 2 in 2cuq
Go back to
Zinc binding site 2 out
of 2 in the Solution Structure of Second Lim Domain From Human Skeletal Muscle Lim-Protein 2
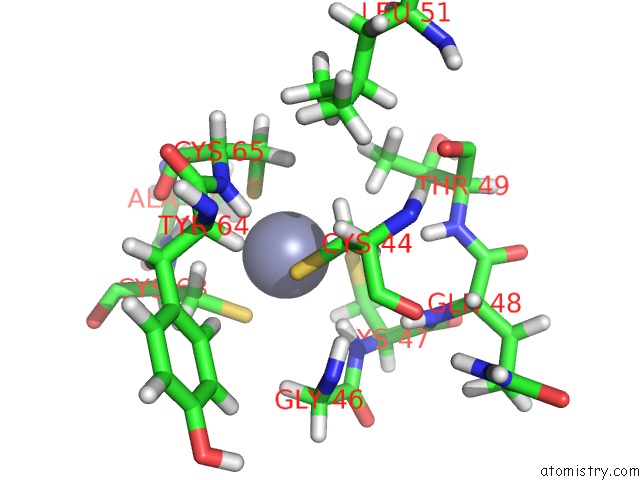
Mono view
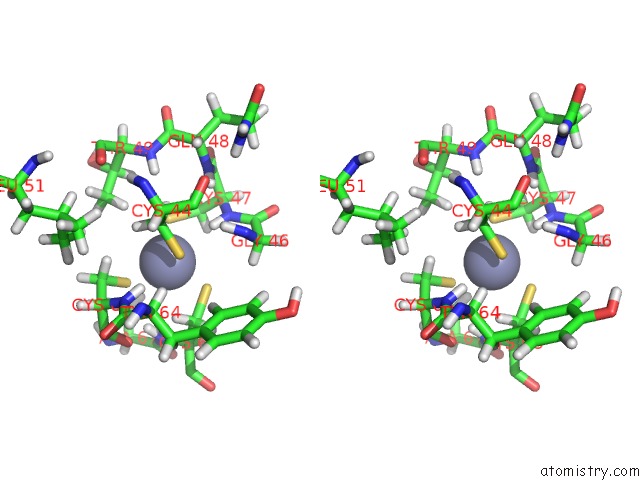
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 2 of Solution Structure of Second Lim Domain From Human Skeletal Muscle Lim-Protein 2 within 5.0Å range:
|
Reference:
T.N.Niraula,
K.Saito,
S.Koshiba,
M.Inoue,
T.Kigawa,
S.Yokoyama.
Solution Structure of Second Lim Domain From Human Skeletal Muscle Lim-Protein 2 To Be Published.
Page generated: Wed Oct 16 22:33:39 2024
Last articles
Fe in 2YXOFe in 2YRS
Fe in 2YXC
Fe in 2YNM
Fe in 2YVJ
Fe in 2YP1
Fe in 2YU2
Fe in 2YU1
Fe in 2YQB
Fe in 2YOO