Zinc »
PDB 1zfp-1zsb »
1zio »
Zinc in PDB 1zio: Phosphotransferase
Enzymatic activity of Phosphotransferase
All present enzymatic activity of Phosphotransferase:
2.7.4.3;
2.7.4.3;
Protein crystallography data
The structure of Phosphotransferase, PDB code: 1zio
was solved by
M.B.Berry,
G.N.Phillips Jr.,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 10.00 / 1.96 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 41.200, 62.165, 41.930, 90.00, 117.10, 90.00 |
| R / Rfree (%) | 16.1 / 22.8 |
Other elements in 1zio:
The structure of Phosphotransferase also contains other interesting chemical elements:
| Magnesium | (Mg) | 1 atom |
Zinc Binding Sites:
The binding sites of Zinc atom in the Phosphotransferase
(pdb code 1zio). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total only one binding site of Zinc was determined in the Phosphotransferase, PDB code: 1zio:
In total only one binding site of Zinc was determined in the Phosphotransferase, PDB code: 1zio:
Zinc binding site 1 out of 1 in 1zio
Go back to
Zinc binding site 1 out
of 1 in the Phosphotransferase
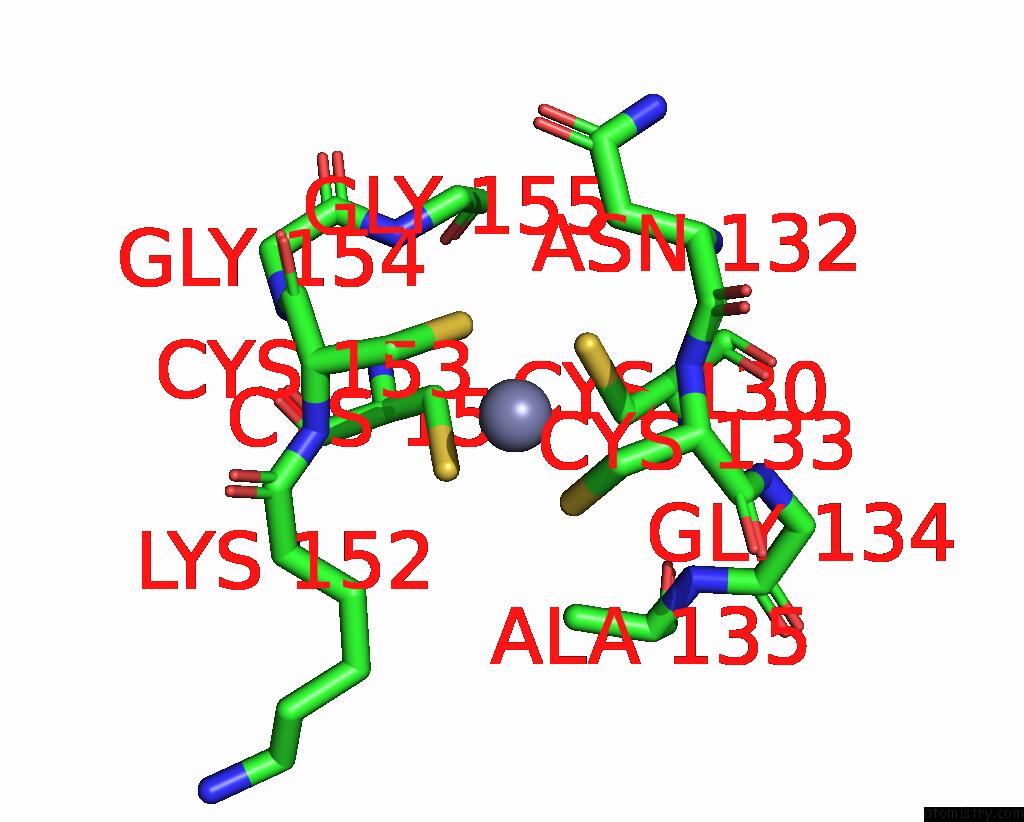
Mono view
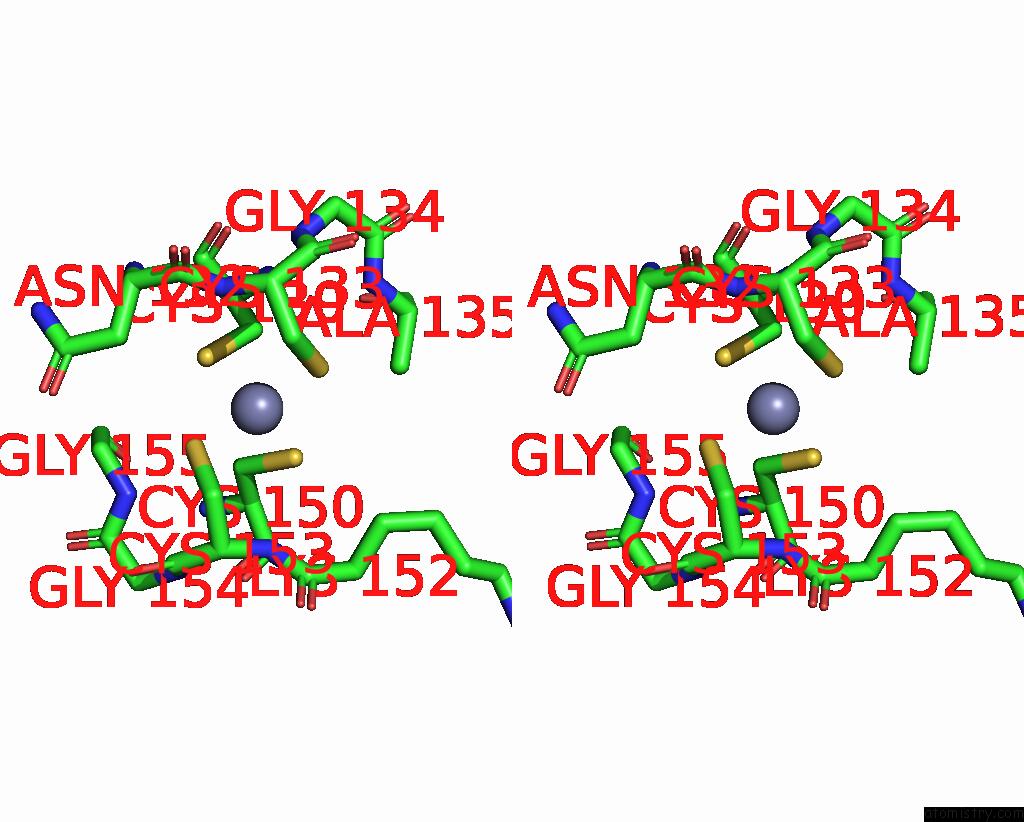
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Phosphotransferase within 5.0Å range:
|
Reference:
M.B.Berry,
G.N.Phillips Jr..
Crystal Structures of Bacillus Stearothermophilus Adenylate Kinase with Bound AP5A, MG2+ AP5A, and MN2+ AP5A Reveal An Intermediate Lid Position and Six Coordinate Octahedral Geometry For Bound MG2+ and MN2+. Proteins V. 32 276 1998.
ISSN: ISSN 0887-3585
PubMed: 9715904
DOI: 10.1002/(SICI)1097-0134(19980815)32:3<276::AID-PROT3>3.0.CO;2-G
Page generated: Wed Oct 16 21:16:14 2024
ISSN: ISSN 0887-3585
PubMed: 9715904
DOI: 10.1002/(SICI)1097-0134(19980815)32:3<276::AID-PROT3>3.0.CO;2-G
Last articles
Ni in 5FPZNi in 5FPX
Ni in 5FM8
Ni in 5FLH
Ni in 5FLE
Ni in 5FJH
Ni in 5FJK
Ni in 5F5I
Ni in 5ERL
Ni in 5FG6