Zinc »
PDB 1z5r-1zfo »
1zat »
Zinc in PDB 1zat: Crystal Structure of An Enterococcus Faecium Peptidoglycan Binding Protein at 2.4 A Resolution
Protein crystallography data
The structure of Crystal Structure of An Enterococcus Faecium Peptidoglycan Binding Protein at 2.4 A Resolution, PDB code: 1zat
was solved by
S.Biarrotte-Sorin,
J.-E.Hugonnet,
J.-L.Mainardi,
L.Gutmann,
L.Rice,
M.Arthur,
C.Mayer,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 21.92 / 2.40 |
| Space group | P 31 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 115.976, 115.976, 68.275, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 22 / 25.7 |
Zinc Binding Sites:
The binding sites of Zinc atom in the Crystal Structure of An Enterococcus Faecium Peptidoglycan Binding Protein at 2.4 A Resolution
(pdb code 1zat). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total only one binding site of Zinc was determined in the Crystal Structure of An Enterococcus Faecium Peptidoglycan Binding Protein at 2.4 A Resolution, PDB code: 1zat:
In total only one binding site of Zinc was determined in the Crystal Structure of An Enterococcus Faecium Peptidoglycan Binding Protein at 2.4 A Resolution, PDB code: 1zat:
Zinc binding site 1 out of 1 in 1zat
Go back to
Zinc binding site 1 out
of 1 in the Crystal Structure of An Enterococcus Faecium Peptidoglycan Binding Protein at 2.4 A Resolution
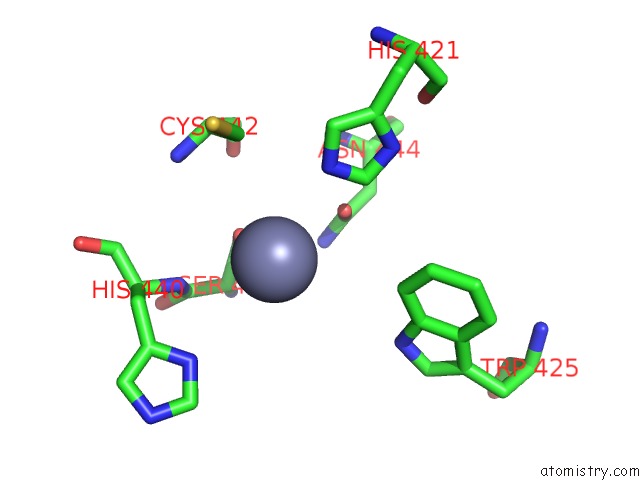
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Crystal Structure of An Enterococcus Faecium Peptidoglycan Binding Protein at 2.4 A Resolution within 5.0Å range:
|
Reference:
S.Biarrotte-Sorin,
J.E.Hugonnet,
V.Delfosse,
J.L.Mainardi,
L.Gutmann,
M.Arthur,
C.Mayer.
Crystal Structure of A Novel Beta-Lactam-Insensitive Peptidoglycan Transpeptidase. J.Mol.Biol. V. 359 533 2006.
ISSN: ISSN 0022-2836
PubMed: 16647082
DOI: 10.1016/J.JMB.2006.03.014
Page generated: Wed Oct 16 21:11:53 2024
ISSN: ISSN 0022-2836
PubMed: 16647082
DOI: 10.1016/J.JMB.2006.03.014
Last articles
Mn in 2BFFMn in 2BFE
Mn in 2BFD
Mn in 2BFC
Mn in 2BFB
Mn in 2BEW
Mn in 2BEV
Mn in 2BEU
Mn in 2BB7
Mn in 2BDX