Zinc »
PDB 1xmo-1xur »
1xtm »
Zinc in PDB 1xtm: Crystal Structure of the Double Mutant Y88H-P104H of A Sod-Like Protein From Bacillus Subtilis.
Protein crystallography data
The structure of Crystal Structure of the Double Mutant Y88H-P104H of A Sod-Like Protein From Bacillus Subtilis., PDB code: 1xtm
was solved by
V.Calderone,
S.Mangani,
L.Banci,
M.Benvenuti,
I.Bertini,
A.Fantoni,
M.S.Viezzoli,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 25.60 / 1.60 |
| Space group | P 21 21 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 52.462, 104.350, 58.756, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 25.3 / 26.6 |
Other elements in 1xtm:
The structure of Crystal Structure of the Double Mutant Y88H-P104H of A Sod-Like Protein From Bacillus Subtilis. also contains other interesting chemical elements:
| Copper | (Cu) | 2 atoms |
Zinc Binding Sites:
The binding sites of Zinc atom in the Crystal Structure of the Double Mutant Y88H-P104H of A Sod-Like Protein From Bacillus Subtilis.
(pdb code 1xtm). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total 5 binding sites of Zinc where determined in the Crystal Structure of the Double Mutant Y88H-P104H of A Sod-Like Protein From Bacillus Subtilis., PDB code: 1xtm:
Jump to Zinc binding site number: 1; 2; 3; 4; 5;
In total 5 binding sites of Zinc where determined in the Crystal Structure of the Double Mutant Y88H-P104H of A Sod-Like Protein From Bacillus Subtilis., PDB code: 1xtm:
Jump to Zinc binding site number: 1; 2; 3; 4; 5;
Zinc binding site 1 out of 5 in 1xtm
Go back to
Zinc binding site 1 out
of 5 in the Crystal Structure of the Double Mutant Y88H-P104H of A Sod-Like Protein From Bacillus Subtilis.
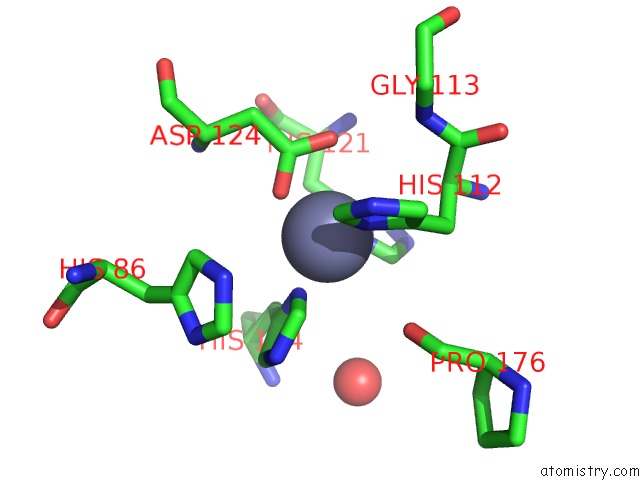
Mono view
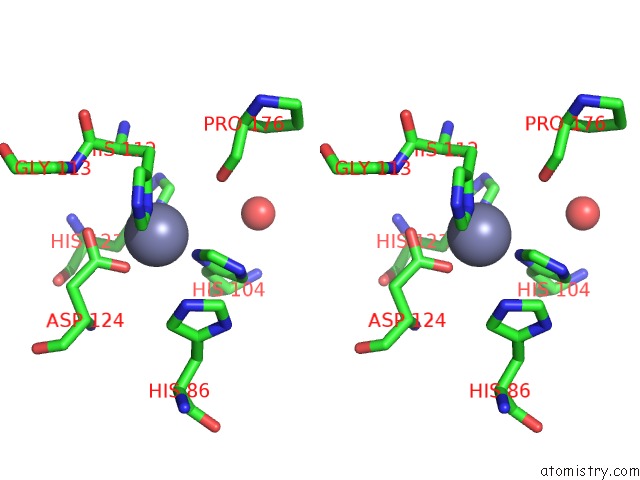
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Crystal Structure of the Double Mutant Y88H-P104H of A Sod-Like Protein From Bacillus Subtilis. within 5.0Å range:
|
Zinc binding site 2 out of 5 in 1xtm
Go back to
Zinc binding site 2 out
of 5 in the Crystal Structure of the Double Mutant Y88H-P104H of A Sod-Like Protein From Bacillus Subtilis.
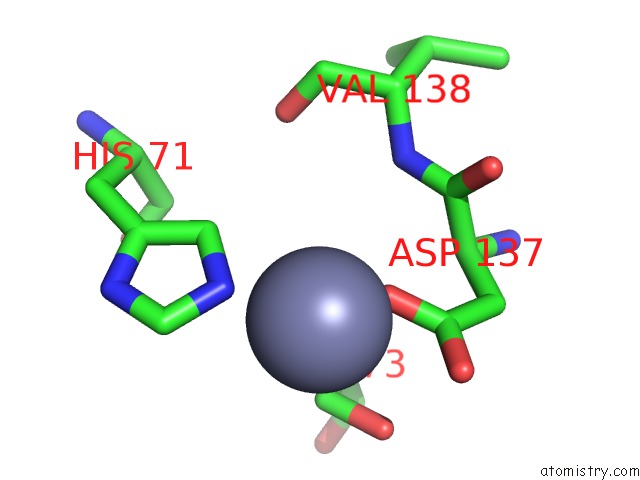
Mono view
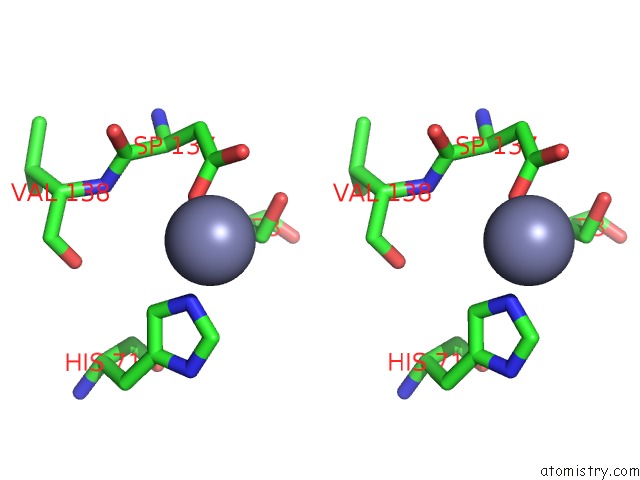
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 2 of Crystal Structure of the Double Mutant Y88H-P104H of A Sod-Like Protein From Bacillus Subtilis. within 5.0Å range:
|
Zinc binding site 3 out of 5 in 1xtm
Go back to
Zinc binding site 3 out
of 5 in the Crystal Structure of the Double Mutant Y88H-P104H of A Sod-Like Protein From Bacillus Subtilis.
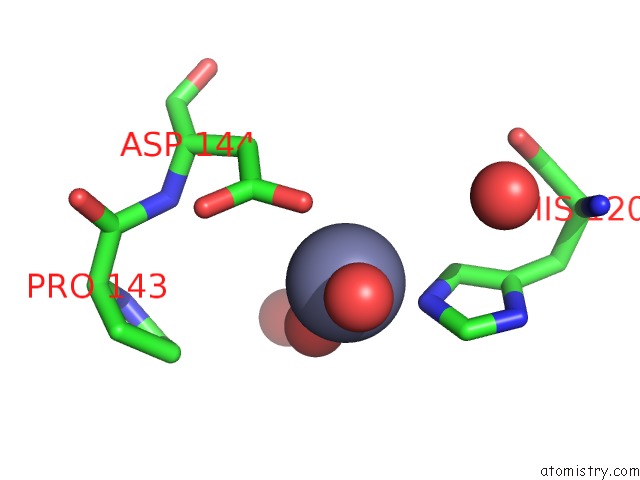
Mono view
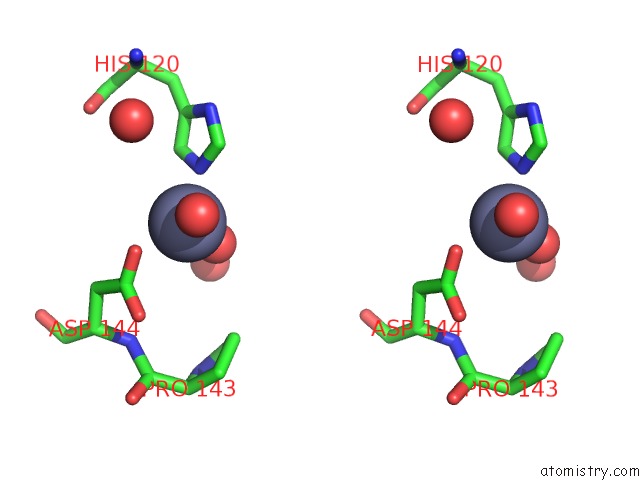
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 3 of Crystal Structure of the Double Mutant Y88H-P104H of A Sod-Like Protein From Bacillus Subtilis. within 5.0Å range:
|
Zinc binding site 4 out of 5 in 1xtm
Go back to
Zinc binding site 4 out
of 5 in the Crystal Structure of the Double Mutant Y88H-P104H of A Sod-Like Protein From Bacillus Subtilis.
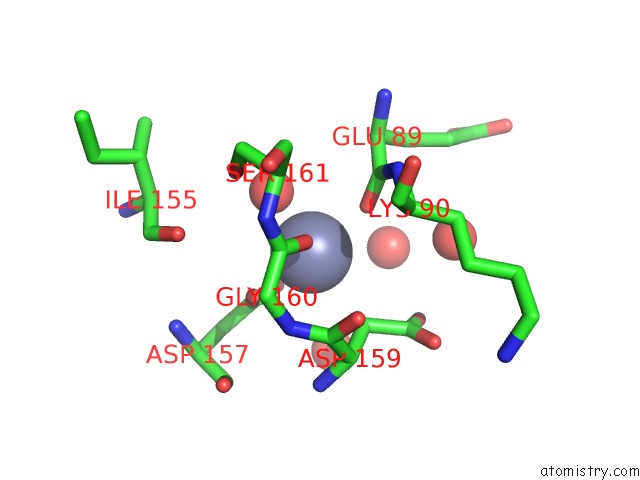
Mono view
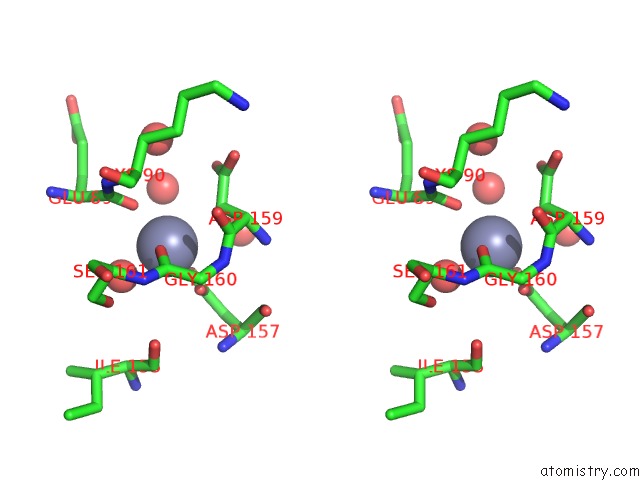
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 4 of Crystal Structure of the Double Mutant Y88H-P104H of A Sod-Like Protein From Bacillus Subtilis. within 5.0Å range:
|
Zinc binding site 5 out of 5 in 1xtm
Go back to
Zinc binding site 5 out
of 5 in the Crystal Structure of the Double Mutant Y88H-P104H of A Sod-Like Protein From Bacillus Subtilis.
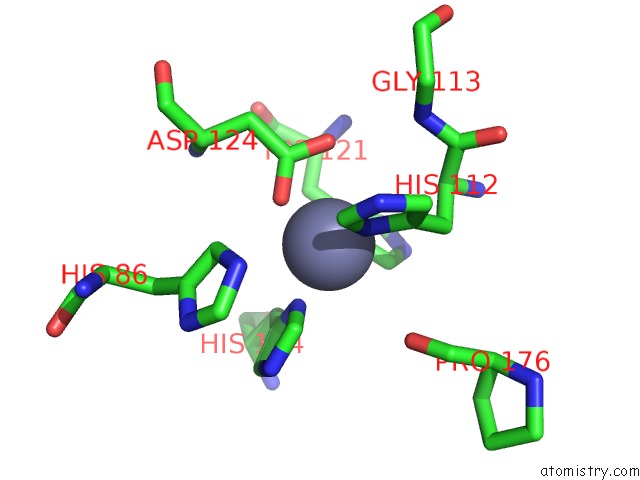
Mono view
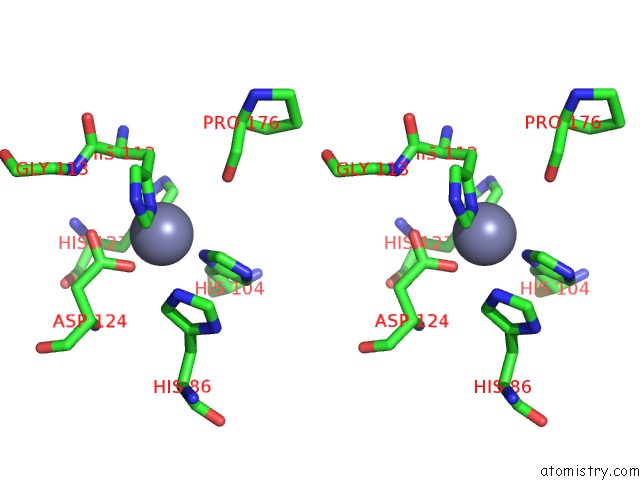
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 5 of Crystal Structure of the Double Mutant Y88H-P104H of A Sod-Like Protein From Bacillus Subtilis. within 5.0Å range:
|
Reference:
L.Banci,
M.Benvenuti,
I.Bertini,
D.E.Cabelli,
V.Calderone,
A.Fantoni,
S.Mangani,
M.Migliardi,
M.S.Viezzoli.
From An Inactive Prokaryotic Sod Homologue to An Active Protein Through Site-Directed Mutagenesis. J.Am.Chem.Soc. V. 127 13287 2005.
ISSN: ISSN 0002-7863
PubMed: 16173759
DOI: 10.1021/JA052790O
Page generated: Wed Oct 16 20:35:43 2024
ISSN: ISSN 0002-7863
PubMed: 16173759
DOI: 10.1021/JA052790O
Last articles
I in 1W51I in 1W50
I in 1V1G
I in 1VKO
I in 1VJ5
I in 1V3P
I in 1V3O
I in 1VAT
I in 1U12
I in 1UTX