Zinc »
PDB 1to5-1u22 »
1tqx »
Zinc in PDB 1tqx: Crystal Structure of PFAL009167 A Putative D-Ribulose 5-Phosphate 3- Epimerase From P.Falciparum
Protein crystallography data
The structure of Crystal Structure of PFAL009167 A Putative D-Ribulose 5-Phosphate 3- Epimerase From P.Falciparum, PDB code: 1tqx
was solved by
J.Caruthers,
J.Bosch,
W.G.J.Hol,
Structural Genomics Of Pathogenicprotozoa Consortium (Sgpp),
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 52.17 / 2.00 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 79.415, 84.454, 120.717, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 21.2 / 23.3 |
Zinc Binding Sites:
The binding sites of Zinc atom in the Crystal Structure of PFAL009167 A Putative D-Ribulose 5-Phosphate 3- Epimerase From P.Falciparum
(pdb code 1tqx). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total 2 binding sites of Zinc where determined in the Crystal Structure of PFAL009167 A Putative D-Ribulose 5-Phosphate 3- Epimerase From P.Falciparum, PDB code: 1tqx:
Jump to Zinc binding site number: 1; 2;
In total 2 binding sites of Zinc where determined in the Crystal Structure of PFAL009167 A Putative D-Ribulose 5-Phosphate 3- Epimerase From P.Falciparum, PDB code: 1tqx:
Jump to Zinc binding site number: 1; 2;
Zinc binding site 1 out of 2 in 1tqx
Go back to
Zinc binding site 1 out
of 2 in the Crystal Structure of PFAL009167 A Putative D-Ribulose 5-Phosphate 3- Epimerase From P.Falciparum
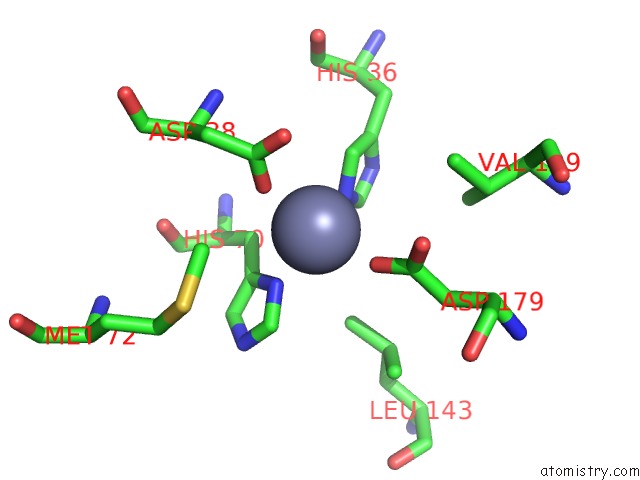
Mono view
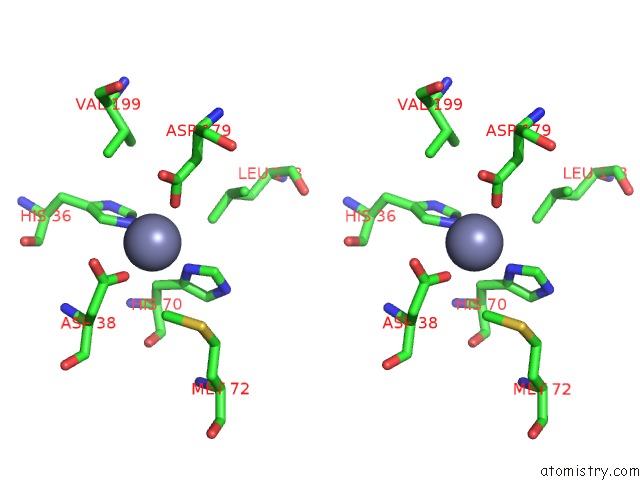
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Crystal Structure of PFAL009167 A Putative D-Ribulose 5-Phosphate 3- Epimerase From P.Falciparum within 5.0Å range:
|
Zinc binding site 2 out of 2 in 1tqx
Go back to
Zinc binding site 2 out
of 2 in the Crystal Structure of PFAL009167 A Putative D-Ribulose 5-Phosphate 3- Epimerase From P.Falciparum
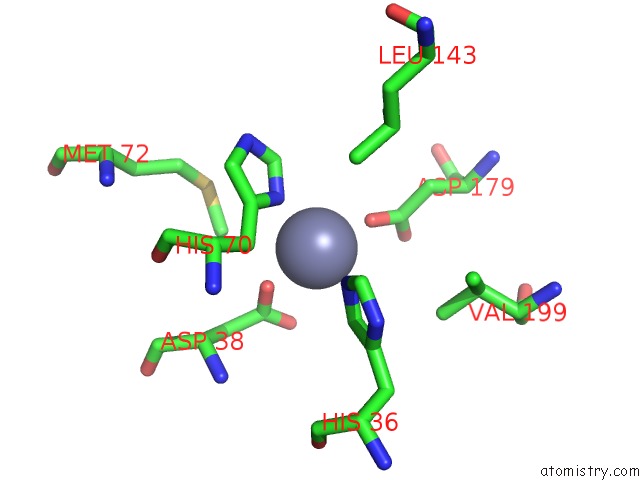
Mono view
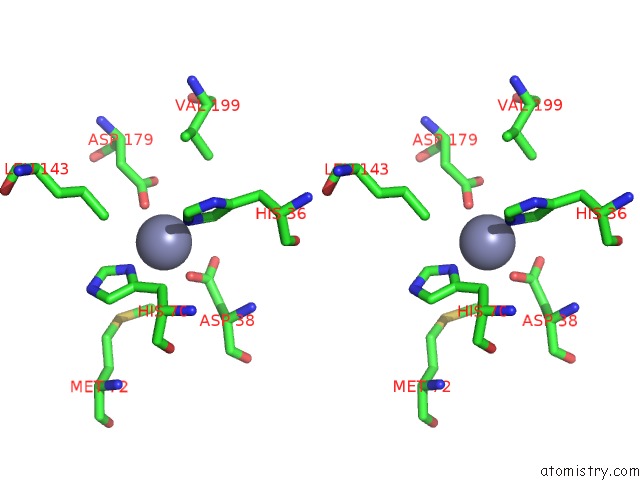
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 2 of Crystal Structure of PFAL009167 A Putative D-Ribulose 5-Phosphate 3- Epimerase From P.Falciparum within 5.0Å range:
|
Reference:
J.Caruthers,
J.Bosch,
F.Buckner,
W.Van Voorhis,
P.Myler,
E.Worthey,
C.Mehlin,
E.Boni,
G.Detitta,
J.Luft,
A.Lauricella,
O.Kalyuzhniy,
L.Anderson,
F.Zucker,
M.Soltis,
W.G.Hol.
Structure of A Ribulose 5-Phosphate 3-Epimerase From Plasmodium Falciparum. Proteins V. 62 338 2006.
ISSN: ISSN 0887-3585
PubMed: 16304640
DOI: 10.1002/PROT.20764
Page generated: Wed Oct 16 19:16:48 2024
ISSN: ISSN 0887-3585
PubMed: 16304640
DOI: 10.1002/PROT.20764
Last articles
F in 6DGTF in 6DFN
F in 6DF6
F in 6DDG
F in 6DDD
F in 6DD4
F in 6DD0
F in 6DBN
F in 6DBM
F in 6DA4