Zinc »
PDB 1rrm-1sdy »
1rtl »
Zinc in PDB 1rtl: Crystal Structure of Hcv NS3 Protease Domain: NS4A Peptide Complex with Covalently Bound Pyrrolidine-5,5-Translactam Inhibitor
Protein crystallography data
The structure of Crystal Structure of Hcv NS3 Protease Domain: NS4A Peptide Complex with Covalently Bound Pyrrolidine-5,5-Translactam Inhibitor, PDB code: 1rtl
was solved by
T.Skarzynski,
D.O.N.Somers,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 20.00 / 2.75 |
| Space group | H 3 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 224.082, 224.082, 75.694, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 18 / 23.4 |
Zinc Binding Sites:
The binding sites of Zinc atom in the Crystal Structure of Hcv NS3 Protease Domain: NS4A Peptide Complex with Covalently Bound Pyrrolidine-5,5-Translactam Inhibitor
(pdb code 1rtl). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total 2 binding sites of Zinc where determined in the Crystal Structure of Hcv NS3 Protease Domain: NS4A Peptide Complex with Covalently Bound Pyrrolidine-5,5-Translactam Inhibitor, PDB code: 1rtl:
Jump to Zinc binding site number: 1; 2;
In total 2 binding sites of Zinc where determined in the Crystal Structure of Hcv NS3 Protease Domain: NS4A Peptide Complex with Covalently Bound Pyrrolidine-5,5-Translactam Inhibitor, PDB code: 1rtl:
Jump to Zinc binding site number: 1; 2;
Zinc binding site 1 out of 2 in 1rtl
Go back to
Zinc binding site 1 out
of 2 in the Crystal Structure of Hcv NS3 Protease Domain: NS4A Peptide Complex with Covalently Bound Pyrrolidine-5,5-Translactam Inhibitor
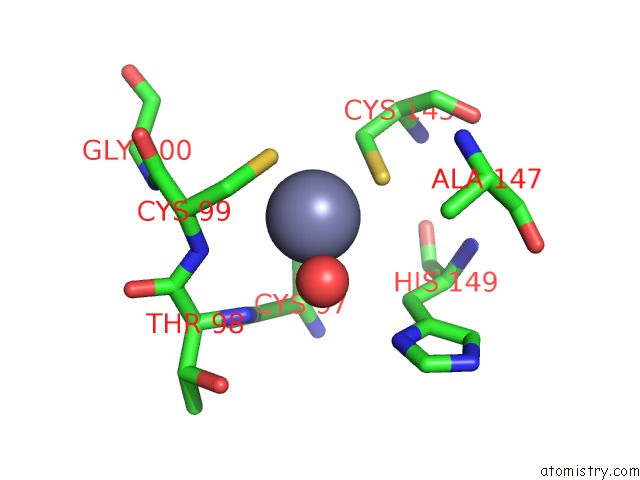
Mono view
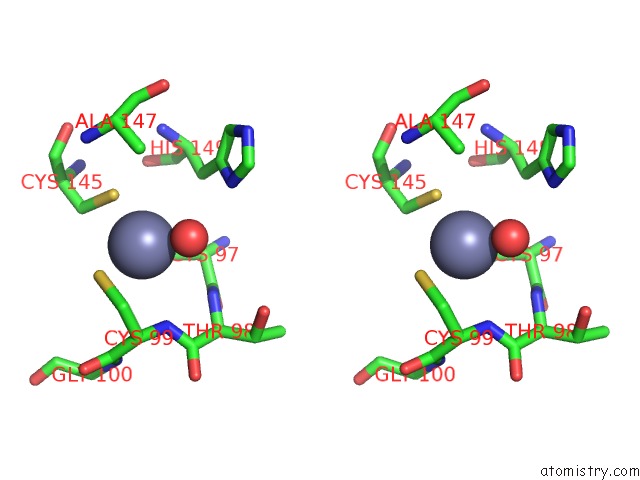
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Crystal Structure of Hcv NS3 Protease Domain: NS4A Peptide Complex with Covalently Bound Pyrrolidine-5,5-Translactam Inhibitor within 5.0Å range:
|
Zinc binding site 2 out of 2 in 1rtl
Go back to
Zinc binding site 2 out
of 2 in the Crystal Structure of Hcv NS3 Protease Domain: NS4A Peptide Complex with Covalently Bound Pyrrolidine-5,5-Translactam Inhibitor
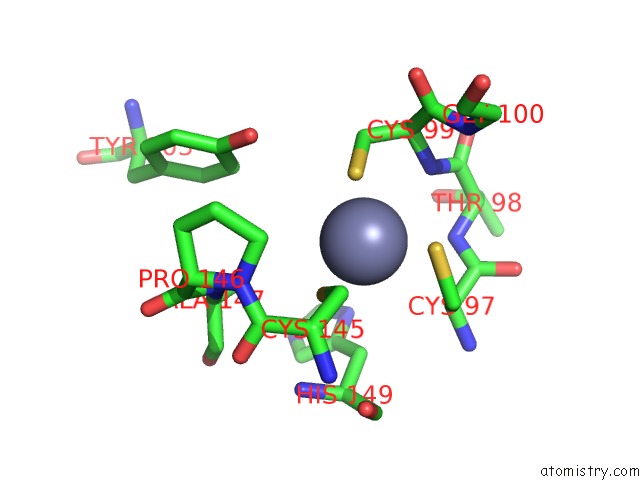
Mono view
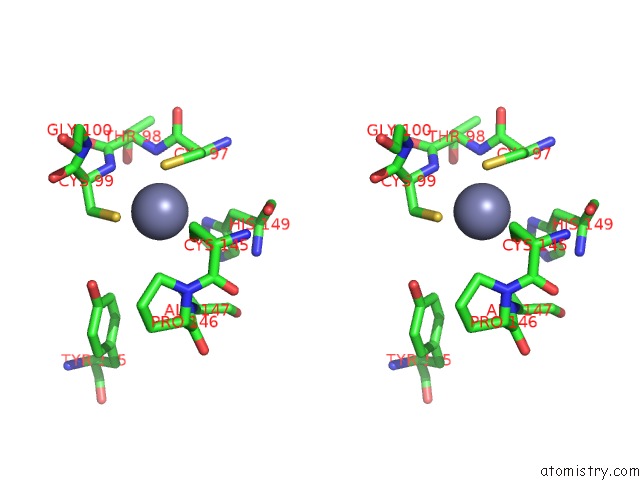
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 2 of Crystal Structure of Hcv NS3 Protease Domain: NS4A Peptide Complex with Covalently Bound Pyrrolidine-5,5-Translactam Inhibitor within 5.0Å range:
|
Reference:
M.J.Slater,
E.M.Amphlett,
D.M.Andrews,
P.Bamborough,
S.J.Carey,
M.R.Johnson,
P.S.Jones,
G.Mills,
N.R.Parry,
D.O.Somers,
A.J.Stewart,
T.Skarzynski.
Pyrrolidine-5,5-Trans-Lactams. 4. Incorporation of A P3/P4 Urea Leads to Potent Intracellular Inhibitors of Hepatitis C Virus NS3/4A Protease Org.Lett. V. 5 4627 2003.
ISSN: ISSN 1523-7060
PubMed: 14627400
DOI: 10.1021/OL035826V
Page generated: Wed Oct 16 18:40:26 2024
ISSN: ISSN 1523-7060
PubMed: 14627400
DOI: 10.1021/OL035826V
Last articles
Fe in 7Z7SFe in 7Z7R
Fe in 7Z0T
Fe in 7Z0S
Fe in 7Z6Q
Fe in 7Z53
Fe in 7Z2L
Fe in 7Z1U
Fe in 7Z10
Fe in 7YZQ