Zinc in PDB 8whu: Spike Trimer of Ba.2.86 in Complex with Two HACE2S
Other elements in 8whu:
The structure of Spike Trimer of Ba.2.86 in Complex with Two HACE2S also contains other interesting chemical elements:
| Chlorine | (Cl) | 2 atoms |
Zinc Binding Sites:
The binding sites of Zinc atom in the Spike Trimer of Ba.2.86 in Complex with Two HACE2S
(pdb code 8whu). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total 2 binding sites of Zinc where determined in the Spike Trimer of Ba.2.86 in Complex with Two HACE2S, PDB code: 8whu:
Jump to Zinc binding site number: 1; 2;
In total 2 binding sites of Zinc where determined in the Spike Trimer of Ba.2.86 in Complex with Two HACE2S, PDB code: 8whu:
Jump to Zinc binding site number: 1; 2;
Zinc binding site 1 out of 2 in 8whu
Go back to
Zinc binding site 1 out
of 2 in the Spike Trimer of Ba.2.86 in Complex with Two HACE2S
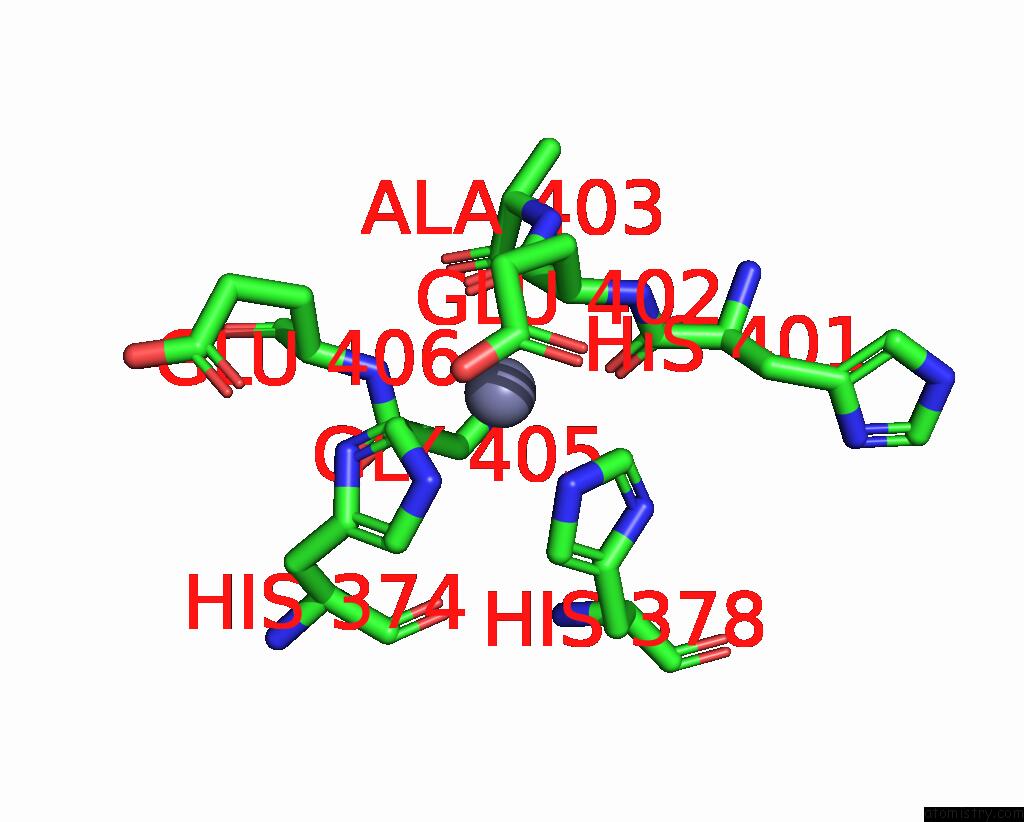
Mono view
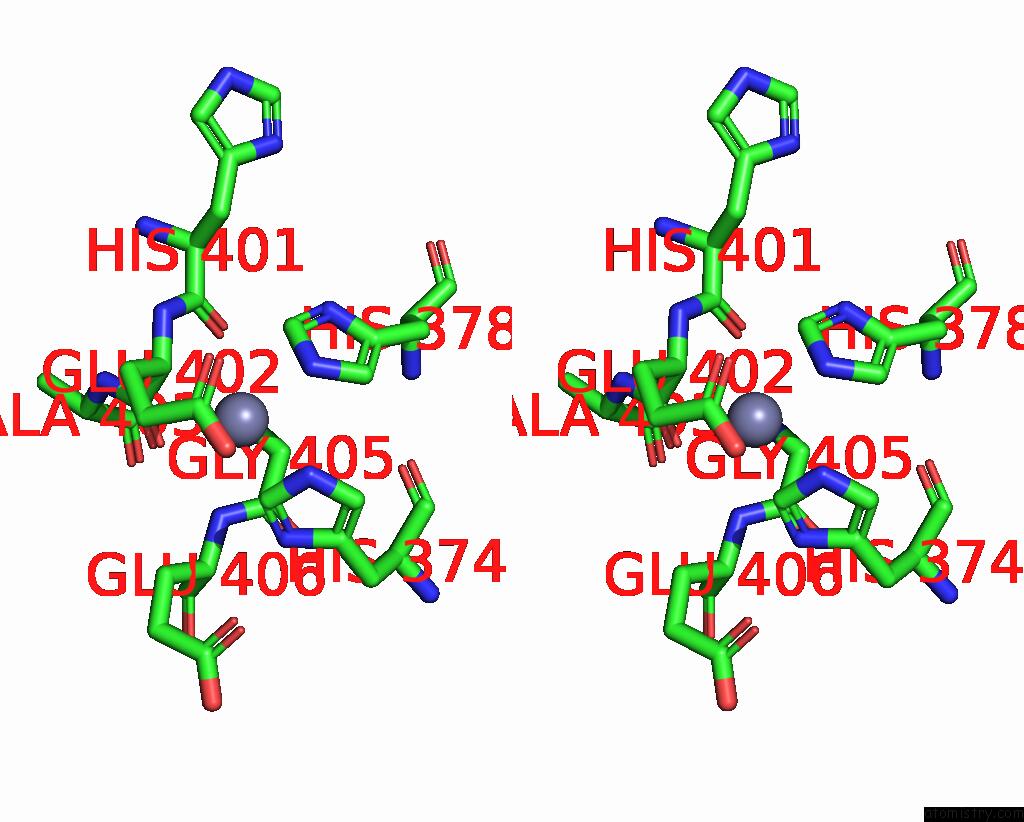
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Spike Trimer of Ba.2.86 in Complex with Two HACE2S within 5.0Å range:
|
Zinc binding site 2 out of 2 in 8whu
Go back to
Zinc binding site 2 out
of 2 in the Spike Trimer of Ba.2.86 in Complex with Two HACE2S
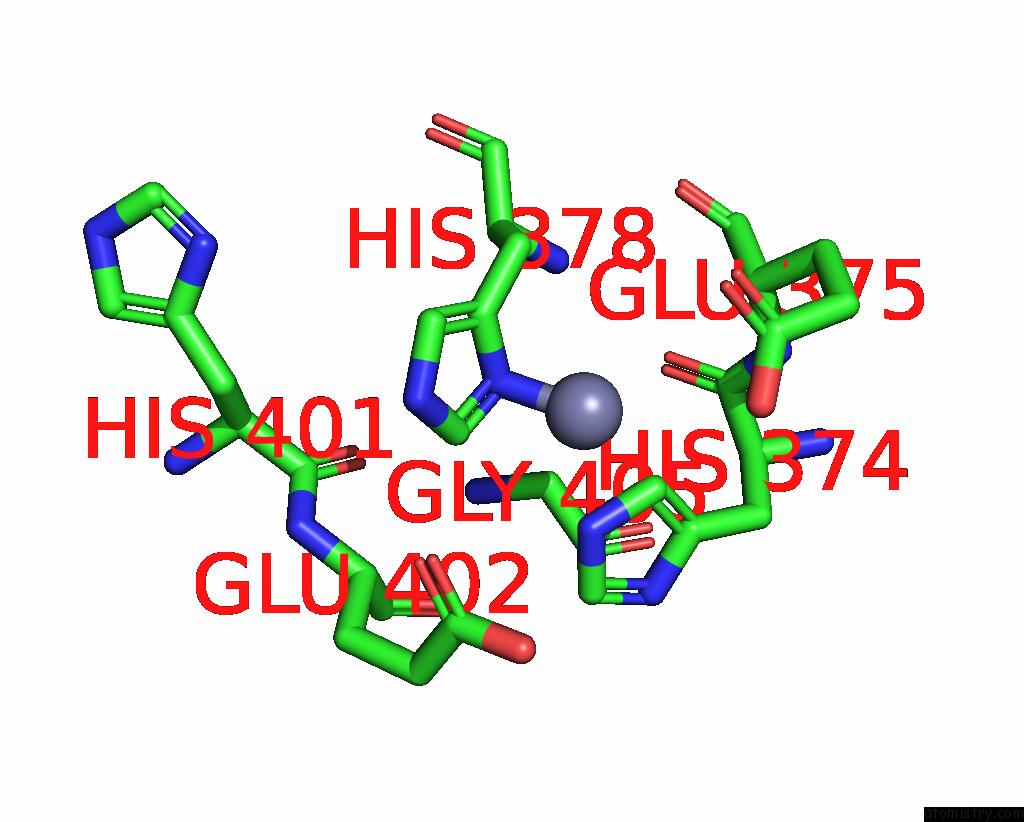
Mono view
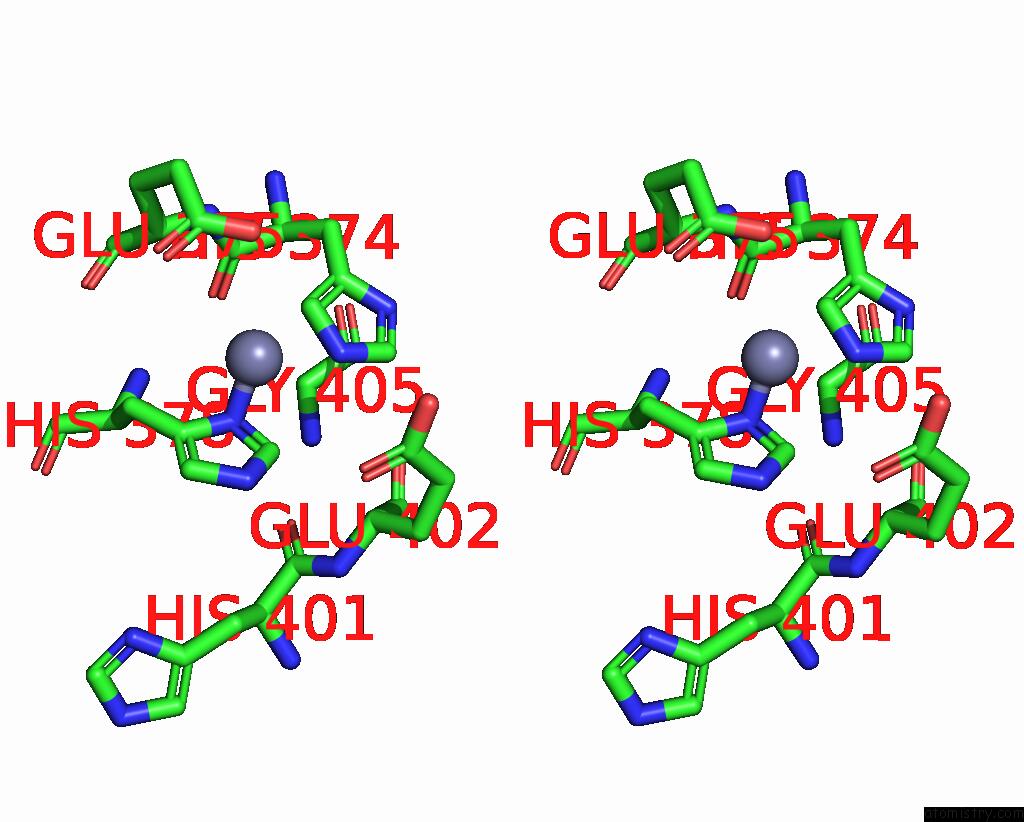
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 2 of Spike Trimer of Ba.2.86 in Complex with Two HACE2S within 5.0Å range:
|
Reference:
P.Liu,
C.Yue,
B.Meng,
T.Xiao,
S.Yang,
S.Liu,
F.Jian,
Q.Zhu,
Y.Yu,
Y.Ren,
P.Wang,
Y.Li,
J.Wang,
X.Mao,
F.Shao,
Y.Wang,
R.K.Gupta,
Y.Cao,
X.Wang.
Spike N354 Glycosylation Augments Sars-Cov-2 Fitness For Human Adaptation Through Structural Plasticity Natl Sci Rev 2024.
ISSN: ESSN 2053-714X
DOI: 10.1093/NSR/NWAE206
Page generated: Thu Oct 31 13:29:27 2024
ISSN: ESSN 2053-714X
DOI: 10.1093/NSR/NWAE206
Last articles
Zn in 9J0NZn in 9J0O
Zn in 9J0P
Zn in 9FJX
Zn in 9EKB
Zn in 9C0F
Zn in 9CAH
Zn in 9CH0
Zn in 9CH3
Zn in 9CH1