Zinc in PDB 7oq4: Cryo-Em Structure of the Atv Rnap Inhibitory Protein (Rip) Bound to the Dna-Binding Channel of the Host'S Rna Polymerase
Enzymatic activity of Cryo-Em Structure of the Atv Rnap Inhibitory Protein (Rip) Bound to the Dna-Binding Channel of the Host'S Rna Polymerase
All present enzymatic activity of Cryo-Em Structure of the Atv Rnap Inhibitory Protein (Rip) Bound to the Dna-Binding Channel of the Host'S Rna Polymerase:
2.7.7.6;
2.7.7.6;
Other elements in 7oq4:
The structure of Cryo-Em Structure of the Atv Rnap Inhibitory Protein (Rip) Bound to the Dna-Binding Channel of the Host'S Rna Polymerase also contains other interesting chemical elements:
| Iron | (Fe) | 3 atoms |
| Magnesium | (Mg) | 1 atom |
Zinc Binding Sites:
The binding sites of Zinc atom in the Cryo-Em Structure of the Atv Rnap Inhibitory Protein (Rip) Bound to the Dna-Binding Channel of the Host'S Rna Polymerase
(pdb code 7oq4). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total 6 binding sites of Zinc where determined in the Cryo-Em Structure of the Atv Rnap Inhibitory Protein (Rip) Bound to the Dna-Binding Channel of the Host'S Rna Polymerase, PDB code: 7oq4:
Jump to Zinc binding site number: 1; 2; 3; 4; 5; 6;
In total 6 binding sites of Zinc where determined in the Cryo-Em Structure of the Atv Rnap Inhibitory Protein (Rip) Bound to the Dna-Binding Channel of the Host'S Rna Polymerase, PDB code: 7oq4:
Jump to Zinc binding site number: 1; 2; 3; 4; 5; 6;
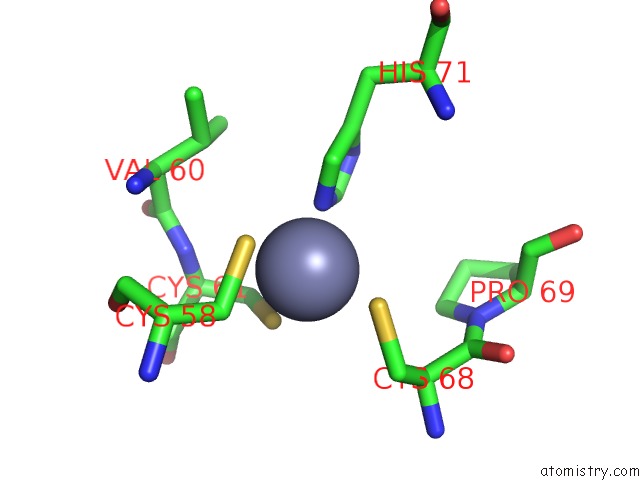
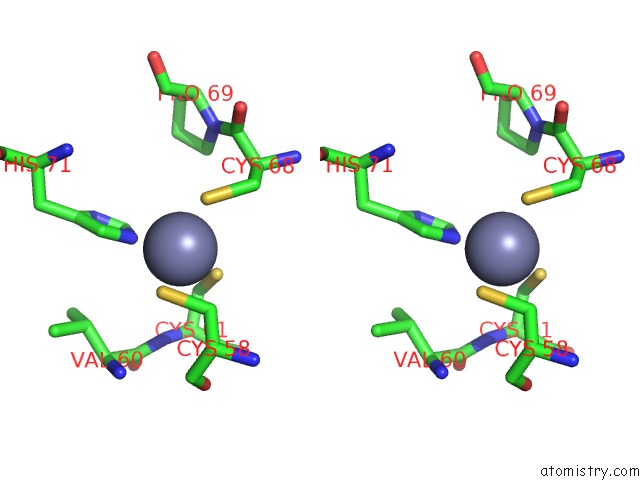
Zinc binding site 1 out of 6 in 7oq4
Go back to
Zinc binding site 1 out
of 6 in the Cryo-Em Structure of the Atv Rnap Inhibitory Protein (Rip) Bound to the Dna-Binding Channel of the Host'S Rna Polymerase
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Cryo-Em Structure of the Atv Rnap Inhibitory Protein (Rip) Bound to the Dna-Binding Channel of the Host'S Rna Polymerase within 5.0Å range:
|
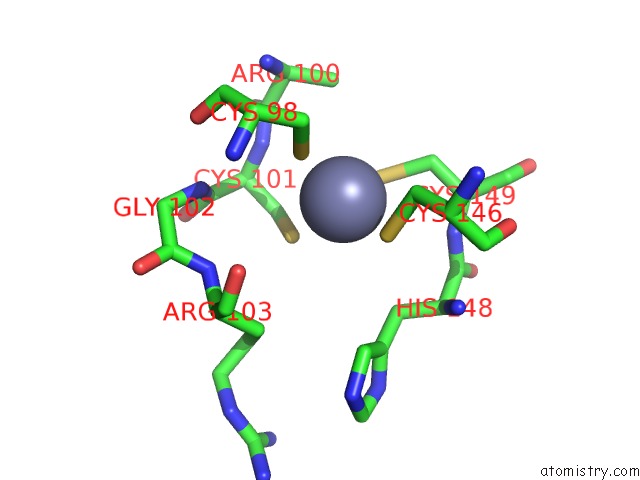
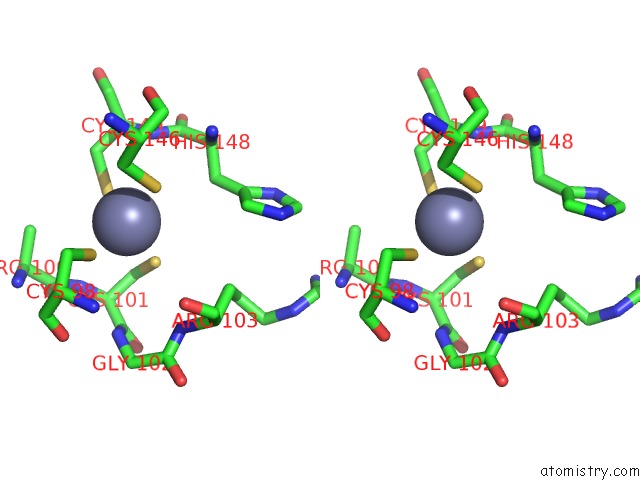
Zinc binding site 2 out of 6 in 7oq4
Go back to
Zinc binding site 2 out
of 6 in the Cryo-Em Structure of the Atv Rnap Inhibitory Protein (Rip) Bound to the Dna-Binding Channel of the Host'S Rna Polymerase
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 2 of Cryo-Em Structure of the Atv Rnap Inhibitory Protein (Rip) Bound to the Dna-Binding Channel of the Host'S Rna Polymerase within 5.0Å range:
|
Zinc binding site 3 out of 6 in 7oq4
Go back to
Zinc binding site 3 out
of 6 in the Cryo-Em Structure of the Atv Rnap Inhibitory Protein (Rip) Bound to the Dna-Binding Channel of the Host'S Rna Polymerase
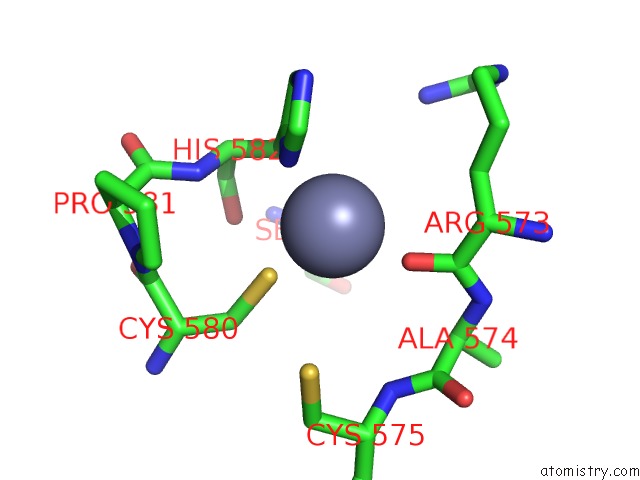
Mono view
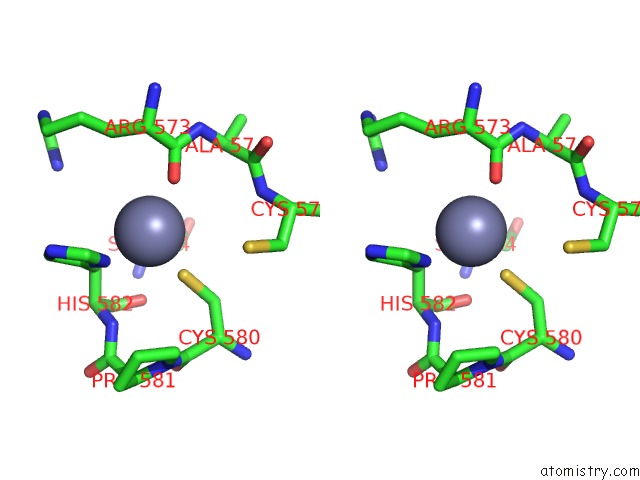
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 3 of Cryo-Em Structure of the Atv Rnap Inhibitory Protein (Rip) Bound to the Dna-Binding Channel of the Host'S Rna Polymerase within 5.0Å range:
|
Zinc binding site 4 out of 6 in 7oq4
Go back to
Zinc binding site 4 out
of 6 in the Cryo-Em Structure of the Atv Rnap Inhibitory Protein (Rip) Bound to the Dna-Binding Channel of the Host'S Rna Polymerase
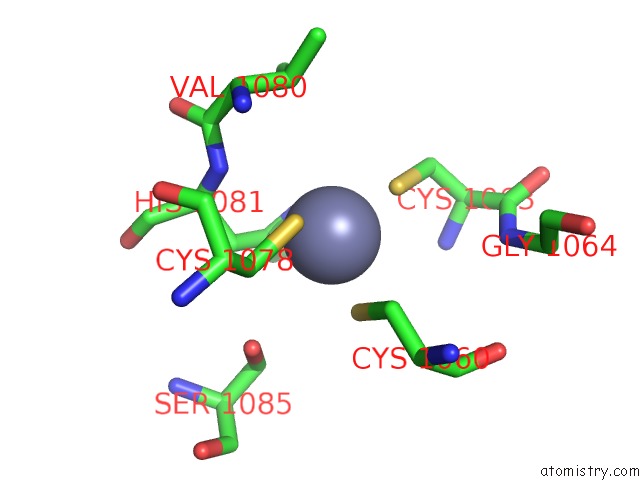
Mono view
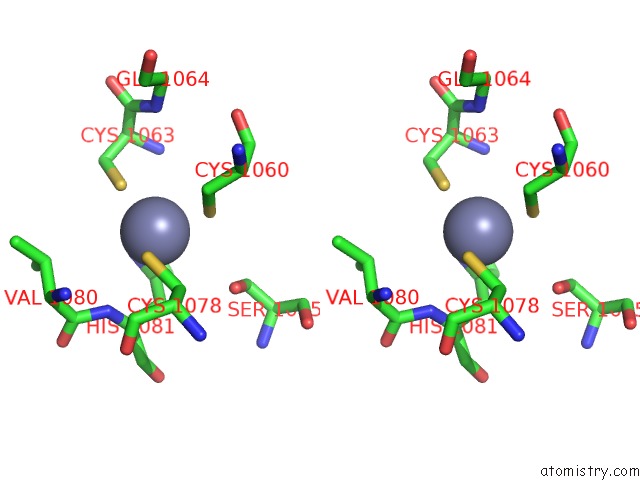
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 4 of Cryo-Em Structure of the Atv Rnap Inhibitory Protein (Rip) Bound to the Dna-Binding Channel of the Host'S Rna Polymerase within 5.0Å range:
|
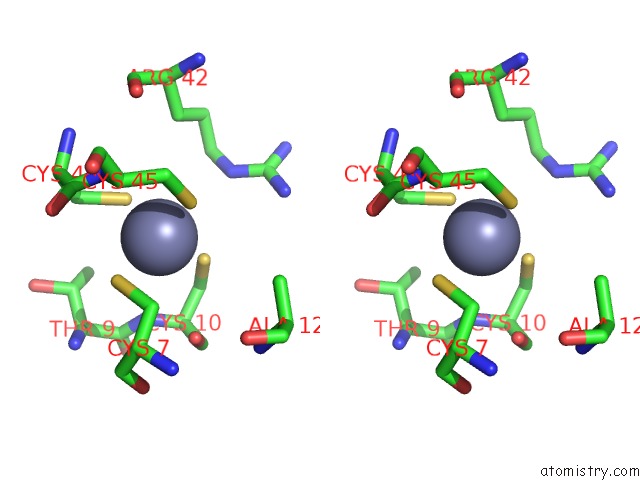
Zinc binding site 5 out of 6 in 7oq4
Go back to
Zinc binding site 5 out
of 6 in the Cryo-Em Structure of the Atv Rnap Inhibitory Protein (Rip) Bound to the Dna-Binding Channel of the Host'S Rna Polymerase
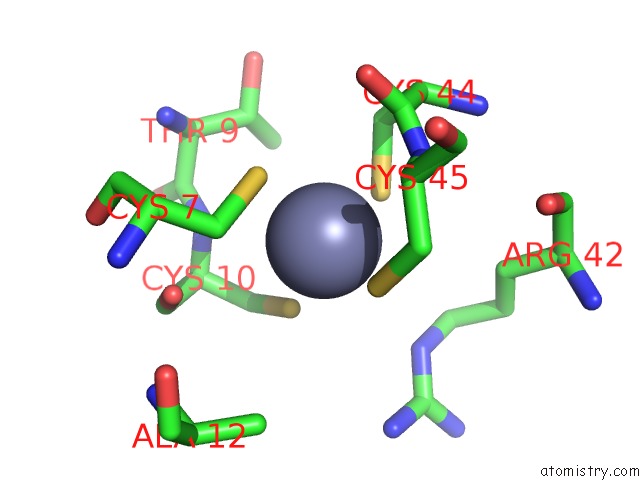
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 5 of Cryo-Em Structure of the Atv Rnap Inhibitory Protein (Rip) Bound to the Dna-Binding Channel of the Host'S Rna Polymerase within 5.0Å range:
|
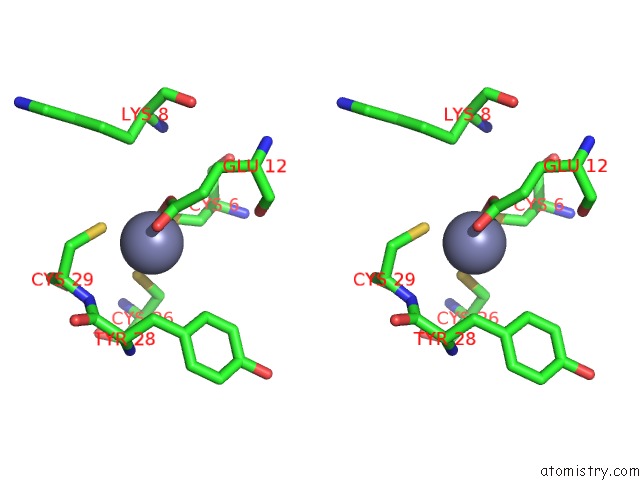
Zinc binding site 6 out of 6 in 7oq4
Go back to
Zinc binding site 6 out
of 6 in the Cryo-Em Structure of the Atv Rnap Inhibitory Protein (Rip) Bound to the Dna-Binding Channel of the Host'S Rna Polymerase
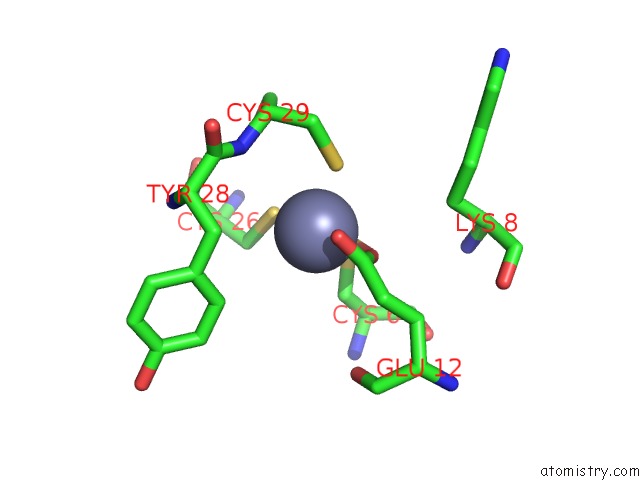
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 6 of Cryo-Em Structure of the Atv Rnap Inhibitory Protein (Rip) Bound to the Dna-Binding Channel of the Host'S Rna Polymerase within 5.0Å range:
|
Reference:
S.Pilotto,
T.Fouqueau,
N.Lukoyanova,
C.Sheppard,
S.Lucas-Staat,
L.M.Diaz-Santin,
D.Matelska,
D.Prangishvili,
A.C.M.Cheung,
F.Werner.
Structural Basis of Rna Polymerase Inhibition By Viral and Host Factors Nat Commun 2021.
ISSN: ESSN 2041-1723
Page generated: Wed Oct 30 08:42:47 2024
ISSN: ESSN 2041-1723
Last articles
Zn in 9J0NZn in 9J0O
Zn in 9J0P
Zn in 9FJX
Zn in 9EKB
Zn in 9C0F
Zn in 9CAH
Zn in 9CH0
Zn in 9CH3
Zn in 9CH1